Characterization of the human submandibular/sublingual saliva glycoproteome using lectin affinity chromatography coupled to multidimensional protein identification technology
- PMID: 21936497
- PMCID: PMC3279195
- DOI: 10.1021/pr200505t
Characterization of the human submandibular/sublingual saliva glycoproteome using lectin affinity chromatography coupled to multidimensional protein identification technology
Abstract
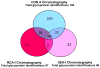
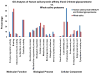
In-depth analysis of the salivary proteome is fundamental to understanding the functions of salivary proteins in the oral cavity and to reveal disease biomarkers involved in different pathophysiological conditions, with the ultimate goal of improving patient diagnosis and prognosis. Submandibular and sublingual glands contribute saliva rich in glycoproteins to the total saliva output, making them valuable sources for glycoproteomic analysis. Lectin-affinity chromatography coupled to mass spectrometry-based shotgun proteomics was used to explore the submandibular/sublingual (SM/SL) saliva glycoproteome. A total of 262 N- and O-linked glycoproteins were identified by multidimensional protein identification technology (MudPIT). Only 38 were previously described in SM and SL salivas from the human salivary N-linked glycoproteome, while 224 were unique. Further comparison analysis with SM/SL saliva of the human saliva proteome, revealed 125 glycoproteins not formerly reported in this secretion. KEGG pathway analyses demonstrated that many of these glycoproteins are involved in processes such as complement and coagulation cascades, cell communication, glycosphingolipid biosynthesis neo-lactoseries, O-glycan biosynthesis, glycan structures-biosynthesis 2, starch and sucrose metabolism, peptidoglycan biosynthesis or others pathways. In summary, lectin-affinity chromatography coupled to MudPIT mass spectrometry identified many novel glycoproteins in SM/SL saliva. These new additions to the salivary proteome may prove to be a critical step for providing reliable biomarkers in the diagnosis of a myriad of oral and systemic diseases.
Figures





References
-
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JR, 3rd, Fisher SJ. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7(5):1994–2006. - PMC - PubMed
-
- Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, Cole S, Fang X, Gonzalez-Begne M, Griffin TJ, Hagen F, Hu S, Wolinsky LE, Lee CS, Malamud D, Melvin JE, Menon R, Mueller M, Qiao R, Rhodus NL, Sevinsky JR, States D, Stephenson JL, Than S, Yates JR, Yu W, Xie H, Xie Y, Omenn GS, Loo JA, Wong DT. Systematic comparison of the human saliva and plasma proteomes. Proteomics: Clin Appl. 2009;3(1):116–34. - PMC - PubMed
-
- Bennick A. Extraoral Functions of Salivary Proteins. J Oral Biosci. 2007;49(1):24–26.
-
- Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. Salivary proteome and its genetic polymorphisms. Ann NY Acad Sci. 2007;1098:22–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

