Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation
- PMID: 21937766
- PMCID: PMC3184980
- DOI: 10.18632/aging.100368
Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation
Abstract
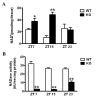
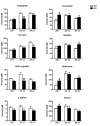
The Intracellular levels of nicotinamide adenine dinucleotide (NAD(+)) are rhythmic and controlled by the circadian clock. However, whether NAD(+) oscillation in turn contributes to circadian physiology is not fully understood. To address this question we analyzed mice mutated for the NAD(+) hydrolase CD38. We found that rhythmicity of NAD(+) was altered in the CD38-deficient mice. The high, chronic levels of NAD(+) results in several anomalies in circadian behavior and metabolism. CD38-null mice display a shortened period length of locomotor activity and alteration in the rest-activity rhythm. Several clock genes and, interestingly, genes involved in amino acid metabolism were deregulated in CD38-null livers. Metabolomic analysis identified alterations in the circadian levels of several amino acids, specifically tryptophan levels were reduced in the CD38-null mice at a circadian time paralleling with elevated NAD(+) levels. Thus, CD38 contributes to behavioral and metabolic circadian rhythms and altered NAD(+) levels influence the circadian clock.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Sahar S, Sassone-Corsi P. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer. 2009;9:886–896. - PubMed
-
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol. Cell. Biol. 2007;8:139–148. - PubMed
-
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. Sirt1 regulates circadian clock gene expression through per2 deacetylation. Cell. 2008;134:317–328. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

