Impaired hepatocellular regeneration in murine sepsis is dependent on regulatory protein levels
- PMID: 21937957
- PMCID: PMC3196846
- DOI: 10.1097/SHK.0b013e31822d60ff
Impaired hepatocellular regeneration in murine sepsis is dependent on regulatory protein levels
Abstract
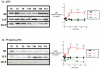
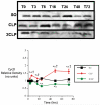
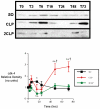
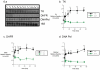
Sepsis is a poorly understood syndrome. Therefore, we examined the mechanisms underlying failed regeneration in sham-operated (SO), mildly septic (cecal ligation and single puncture [CLP]), and severely septic (cecal ligation with two punctures [2CLP]) C57Bl6 mice. Relative to no operation (T0) or SO, CLP, but not 2CLP, increased the number of cells staining for proliferating cell nuclear antigen, a marker for cell division. Levels of the retinoblastoma protein (pRb) were detected at T0 and after SO. CLP increased pRb abundance, whereas 2CLP decreased it. Changes in phosphorylated pRb were similar but more profound. The abundance of the transcription factor E2F was unaltered by SO, CLP, or 2CLP. However, E2F DNA binding activity, although unchanged after SO, increased after CLP and decreased after 2CLP. The abundance of cyclin D1 in nuclear fractions increased following CLP but decreased after 2CLP. Neither SO nor 2CLP altered the abundance of the cyclin-dependent kinase (cdk) 4. However, cdk-4 abundance increased after CLP. Finally, CLP increased the steady-state abundance of the mRNAs encoding thymidine kinase, DNA polymerase α, and dihydrofolate reductase, all required for DNA replication. No changes were noted after 2CLP. We conclude that 2CLP impaired hepatocyte proliferation following 2CLP in part via impaired cyclin D1/cdk-4-induced phosphorylation of pRb, maintaining the association between pRb and E2F and inhibited E2F transcriptional activity.
Figures



References
-
- Angus DC, Linde-Zwerbel WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: Analysis of incidence, outcome and associated costs of care. Crit Care Med. 2001;29:1303–1310. - PubMed
-
- Kohl BA, Deutschman CS. The inflammatory response in organ injury. In: Newman MF, Fleisher LA, Fink MP, editors. Perioperative Management. Saunders/Elsevier; Philadelphia: 2008. pp. 19–28.
-
- Rubulotta F, Marshall JC, Ramsay G, Nelson D, Levy M, Williams M. Predisposition, insult/infection, response, and organ dysfunction: A new model for staging severe sepsis. Crit Care Med. 2009;37:1329–1335. - PubMed
-
- Andrejko KM, Deutschman CS. Altered hepatic gene expression in fecal peritonitis: changes in transcription of gluconeogenic, beta-oxidative, and ureagenic genes. Shock. 1997;7:164–169. - PubMed
-
- Deutschman CS, Cereda M, Ochroch EA, Raj NR. Sepsis-induced cholestasis, steatosis, hepatocellular injury, and impaired hepatocellular regeneration are enhanced in interleukin-6 −/− mice. Crit Care Med. 2006;34:2613–2620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

