Bioinformatics and systems biology of the lipidome
- PMID: 21939287
- PMCID: PMC3383319
- DOI: 10.1021/cr200295k
Bioinformatics and systems biology of the lipidome
Figures














References
-
- Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman GT, Reichart D, Merrill AH, Sullards MC, Wang E, Murphy RC, Raetz CR, Garrett T, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. J. Biol. Chem. 2010;285:39976. - PMC - PubMed
-
- Wymann MP, Schneiter R. Nat. Rev. Mol. Cell Biol. 2008;9:162. - PubMed
-
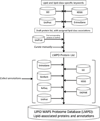
- LIPID MAPS - Nature Lipidomics Gateway. www.lipidmaps.org. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

