Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy
- PMID: 21939668
- PMCID: PMC3220764
- DOI: 10.1016/j.jmb.2011.09.008
Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy
Abstract
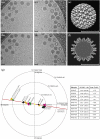
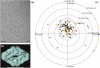
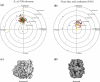
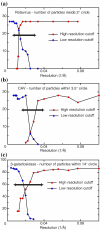
The comparison of a pair of electron microscope images recorded at different specimen tilt angles provides a powerful approach for evaluating the quality of images, image-processing procedures, or three-dimensional structures. Here, we analyze tilt-pair images recorded from a range of specimens with different symmetries and molecular masses and show how the analysis can produce valuable information not easily obtained otherwise. We show that the accuracy of orientation determination of individual single particles depends on molecular mass, as expected theoretically since the information in each particle image increases with molecular mass. The angular uncertainty is less than 1° for particles of high molecular mass (~50 MDa), several degrees for particles in the range 1-5 MDa, and tens of degrees for particles below 1 MDa. Orientational uncertainty may be the major contributor to the effective temperature factor (B-factor) describing contrast loss and therefore the maximum resolution of a structure determination. We also made two unexpected observations. Single particles that are known to be flexible showed a wider spread in orientation accuracy, and the orientations of the largest particles examined changed by several degrees during typical low-dose exposures. Smaller particles presumably also reorient during the exposure; hence, specimen movement is a second major factor that limits resolution. Tilt pairs thus enable assessment of orientation accuracy, map quality, specimen motion, and conformational heterogeneity. A convincing tilt-pair parameter plot, where 60% of the particles show a single cluster around the expected tilt axis and tilt angle, provides confidence in a structure determined using electron cryomicroscopy.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


References
-
- DeRosier D.J., Klug A. Reconstruction of three dimensional structures from electron micrographs. Nature. 1968;217:130–134. - PubMed
-
- Crowther R.A., Amos L.A., Finch J.T., De Rosier D.J., Klug A. Three dimensional reconstructions of spherical viruses by Fourier synthesis from electron micrographs. Nature. 1970;226:421–425. - PubMed
-
- Henderson R., Unwin P.N.T. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. - PubMed
-
- Dubochet J., Adrian M., Chang J.J., Homo J.C., Lepault J., McDowall A.W., Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

