BitterDB: a database of bitter compounds
- PMID: 21940398
- PMCID: PMC3245057
- DOI: 10.1093/nar/gkr755
BitterDB: a database of bitter compounds
Abstract
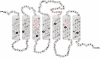
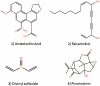
Basic taste qualities like sour, salty, sweet, bitter and umami serve specific functions in identifying food components found in the diet of humans and animals, and are recognized by proteins in the oral cavity. Recognition of bitter taste and aversion to it are thought to protect the organism against the ingestion of poisonous food compounds, which are often bitter. Interestingly, bitter taste receptors are expressed not only in the mouth but also in extraoral tissues, such as the gastrointestinal tract, indicating that they may play a role in digestive and metabolic processes. BitterDB database, available at http://bitterdb.agri.huji.ac.il/bitterdb/, includes over 550 compounds that were reported to taste bitter to humans. The compounds can be searched by name, chemical structure, similarity to other bitter compounds, association with a particular human bitter taste receptor, and so on. The database also contains information on mutations in bitter taste receptors that were shown to influence receptor activation by bitter compounds. The aim of BitterDB is to facilitate studying the chemical features associated with bitterness. These studies may contribute to predicting bitterness of unknown compounds, predicting ligands for bitter receptors from different species and rational design of bitterness modulators.
Figures

References
-
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. - PubMed
-
- Drewnowski A, Gomez-Carneros C. Bitter taste, phytonutrients, and the consumer: a review. Am J. Clin. Nutr. 2000;72:1424–1435. - PubMed
-
- Hofmann T. Identification of the key bitter compounds in our daily diet is a prerequisite for the understanding of the hTAS2R gene polymorphisms affecting food choice. Ann N Y Acad. Sci. 2009;1170:116–125. - PubMed
-
- Rodgers S, Busch J, Peters H, Christ-Hazelhof E. Building a tree of knowledge: analysis of bitter molecules. Chem. Senses. 2005;30:547–557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

