A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript
- PMID: 21940503
- PMCID: PMC3189046
- DOI: 10.1073/pnas.1110904108
A nucleolar protein, H19 opposite tumor suppressor (HOTS), is a tumor growth inhibitor encoded by a human imprinted H19 antisense transcript
Abstract
The H19 gene, which localizes within a chromosomal region on human chromosome 11p15 that is commonly lost in Wilms tumor (WT), encodes an imprinted untranslated RNA. However, the biological significance of the H19 noncoding transcript remains unresolved because replacement of the RNA transcript with a neocassette has no obvious phenotypic effect. Here we show that the human H19 locus also encodes a maternally expressed, translated gene, antisense to the known H19 transcript, which is conserved in primates. This gene, termed HOTS for H19 opposite tumor suppressor, encodes a protein that localizes to the nucleus and nucleolus and that interacts with the human enhancer of rudimentary homolog (ERH) protein. WTs that show loss of heterozygosity of 11p15 or loss of imprinting of IGF2 also silence HOTS (7/7 and 10/10, respectively). Overexpression of HOTS inhibits Wilms, rhabdoid, rhabdomyosarcoma, and choriocarcinoma tumor cell growth, and silencing HOTS by RNAi increases in vitro colony formation and in vivo tumor growth. These results demonstrate that the human H19 locus harbors an imprinted gene encoding a tumor suppressor protein within the long-sought WT2 locus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

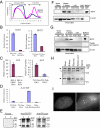

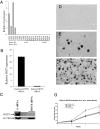
Comment in
-
Tumour suppressors: HOTS makes antisense.Nat Rev Cancer. 2011 Oct 7;11(11):758. doi: 10.1038/nrc3154. Nat Rev Cancer. 2011. PMID: 21979306 No abstract available.
References
-
- Arney KL. H19 and Igf2: Enhancing the confusion? Trends Genet. 2003;19:17–23. - PubMed
-
- Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. - PubMed
-
- Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

