Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs
- PMID: 21940797
- PMCID: PMC3178455
- DOI: 10.1242/jcs.087692
Nuclear relocalisation of cytoplasmic poly(A)-binding proteins PABP1 and PABP4 in response to UV irradiation reveals mRNA-dependent export of metazoan PABPs
Abstract
Poly(A)-binding protein 1 (PABP1) has a fundamental role in the regulation of mRNA translation and stability, both of which are crucial for a wide variety of cellular processes. Although generally a diffuse cytoplasmic protein, it can be found in discrete foci such as stress and neuronal granules. Mammals encode several additional cytoplasmic PABPs that remain poorly characterised, and with the exception of PABP4, appear to be restricted in their expression to a small number of cell types. We have found that PABP4, similarly to PABP1, is a diffusely cytoplasmic protein that can be localised to stress granules. However, UV exposure unexpectedly relocalised both proteins to the nucleus. Nuclear relocalisation of PABPs was accompanied by a reduction in protein synthesis but was not linked to apoptosis. In examining the mechanism of PABP relocalisation, we found that it was related to a change in the distribution of poly(A) RNA within cells. Further investigation revealed that this change in RNA distribution was not affected by PABP knockdown but that perturbations that block mRNA export recapitulate PABP relocalisation. Our results support a model in which nuclear export of PABPs is dependent on ongoing mRNA export, and that a block in this process following UV exposure leads to accumulation of cytoplasmic PABPs in the nucleus. These data also provide mechanistic insight into reports that transcriptional inhibitors and expression of certain viral proteins cause relocation of PABP to the nucleus.
@ 2011. Published by The Company of Biologists Ltd
Figures
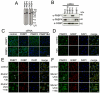

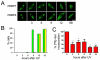

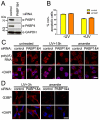

References
-
- Afonina E., Stauber R., Pavlakis G. N. (1998). The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273, 13015-13021 - PubMed
-
- Albrecht M., Lengauer T. (2004). Survey on the PABC recognition motif PAM2. Biochem. Biophys. Res. Commun. 316, 129-138 - PubMed
-
- Anderson P., Kedersha N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430-436 - PubMed
-
- Batista L. F., Kaina B., Meneghini R., Menck C. F. (2009). How DNA lesions are turned into powerful killing structures: insights from UV-induced apoptosis. Mutat. Res. 681, 197-208 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

