Biological functions of p53 isoforms through evolution: lessons from animal and cellular models
- PMID: 21941372
- PMCID: PMC3214904
- DOI: 10.1038/cdd.2011.120
Biological functions of p53 isoforms through evolution: lessons from animal and cellular models
Abstract
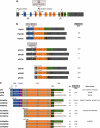
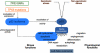
The TP53 tumour-suppressor gene is expressed as several protein isoforms generated by different mechanisms, including use of alternative promoters, splicing sites and translational initiation sites, that are conserved through evolution and within the TP53 homologues, TP63 and TP73. Although first described in the eighties, the importance of p53 isoforms in regulating the suppressive functions of p53 has only become evident in the last 10 years, by analogy with observations that p63 and p73 isoforms appeared indispensable to fully understand the biological functions of TP63 and TP73. This review summarizes recent advances in the field of 'p53 isoforms', including new data on p63 and p73 isoforms. Details of the alternative mechanisms that produce p53 isoforms and cis- and trans-regulators identified are provided. The main focus is on their biological functions (apoptosis, cell cycle, aging and so on) in cellular and animal models, including mouse, zebrafish and Drosophila. Finally, the deregulation of p53 isoform expression in human cancers is reviewed. Based on these latest results, several developments are expected in the future: the identification of drugs modulating p53 isoform expression; the generation of animal models and the evaluation of the use of p53 isoform as biomarkers in human cancers.
Figures

References
-
- Levine AJ, Tomasini R, McKeon FD, Mak TW, Melino G. The p53 family: guardians of maternal reproduction. Nat Rev Mol Cell Biol. 2011;12:259–265. - PubMed
-
- Vilborg A, Wilhelm MT, Wiman KG. Regulation of tumor suppressor p53 at the RNA level. J Mol Med. 2010;88:645–652. - PubMed
-
- Garritano S, Gemignani F, Palmero EI, Olivier M, Martel-Planche G, Le Calvez-Kelm F, et al. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: evidence for a founder effect. Hum Mutat. 2010;31:143–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

