Nucleosome-coupled expression differences in closely-related species
- PMID: 21942931
- PMCID: PMC3209474
- DOI: 10.1186/1471-2164-12-466
Nucleosome-coupled expression differences in closely-related species
Abstract
Background: Genome-wide nucleosome occupancy is negatively related to the average level of transcription factor motif binding based on studies in yeast and several other model organisms. The degree to which nucleosome-motif interactions relate to phenotypic changes across species is, however, unknown.
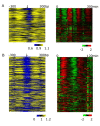
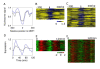
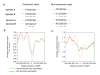
Results: We address this challenge by generating nucleosome positioning and cell cycle expression data for Saccharomyces bayanus and show that differences in nucleosome occupancy reflect cell cycle expression divergence between two yeast species, S. bayanus and S. cerevisiae. Specifically, genes with nucleosome-depleted MBP1 motifs upstream of their coding sequence show periodic expression during the cell cycle, whereas genes with nucleosome-shielded motifs do not. In addition, conserved cell cycle regulatory motifs across these two species are more nucleosome-depleted compared to those that are not conserved, suggesting that the degree of conservation of regulatory sites varies, and is reflected by nucleosome occupancy patterns. Finally, many changes in cell cycle gene expression patterns across species can be correlated to changes in nucleosome occupancy on motifs (rather than to the presence or absence of motifs).
Conclusions: Our observations suggest that alteration of nucleosome occupancy is a previously uncharacterized feature related to the divergence of cell cycle expression between species.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

