How yeast re-programmes its transcriptional profile in response to different nutrient impulses
- PMID: 21943358
- PMCID: PMC3224505
- DOI: 10.1186/1752-0509-5-148
How yeast re-programmes its transcriptional profile in response to different nutrient impulses
Abstract
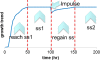
Background: A microorganism is able to adapt to changes in its physicochemical or nutritional environment and this is crucial for its survival. The yeast, Saccharomyces cerevisiae, has developed mechanisms to respond to such environmental changes in a rapid and effective manner; such responses may demand a widespread re-programming of gene activity. The dynamics of the re-organization of the cellular activities of S. cerevisiae in response to the sudden and transient removal of either carbon or nitrogen limitation has been studied by following both the short- and long-term changes in yeast's transcriptomic profiles.
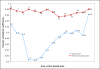
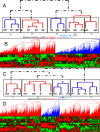
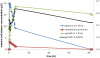
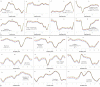
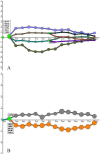
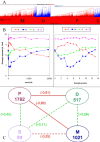
Results: The study, which spans timescales from seconds to hours, has revealed the hierarchy of metabolic and genetic regulatory switches that allow yeast to adapt to, and recover from, a pulse of a previously limiting nutrient. At the transcriptome level, a glucose impulse evoked significant changes in the expression of genes concerned with glycolysis, carboxylic acid metabolism, oxidative phosphorylation, and nucleic acid and sulphur metabolism. In ammonium-limited cultures, an ammonium impulse resulted in the significant changes in the expression of genes involved in nitrogen metabolism and ion transport. Although both perturbations evoked significant changes in the expression of genes involved in the machinery and process of protein synthesis, the transcriptomic response was delayed and less complex in the case of an ammonium impulse. Analysis of the regulatory events by two different system-level, network-based approaches provided further information about dynamic organization of yeast cells as a response to a nutritional change.
Conclusions: The study provided important information on the temporal organization of transcriptomic organization and underlying regulatory events as a response to both carbon and nitrogen impulse. It has also revealed the importance of a long-term dynamic analysis of the response to the relaxation of a nutritional limitation to understand the molecular basis of the cells' dynamic behaviour.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

