Whole-exome sequencing of DNA from peripheral blood mononuclear cells (PBMC) and EBV-transformed lymphocytes from the same donor
- PMID: 21943378
- PMCID: PMC3203102
- DOI: 10.1186/1471-2164-12-464
Whole-exome sequencing of DNA from peripheral blood mononuclear cells (PBMC) and EBV-transformed lymphocytes from the same donor
Abstract
Background: The creation of lymphoblastoid cell lines (LCLs) through Epstein-Barr virus (EBV) transformation of B-lymphocytes can result in a valuable biomaterial for cell biology research and a renewable source of DNA. While LCLs have been used extensively in cellular and genetic studies, the process of cell transformation and expansion during culturing may introduce genomic changes that may impact their use and the interpretation of subsequent genetic findings.
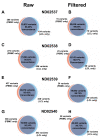
Results: We performed whole exome sequencing on a tetrad family using DNA derived from peripheral blood mononuclear cells (PBMCs) and LCLs from each individual. We generated over 4.7 GB of mappable sequence to a 125X read coverage per sample. An average of 19,354 genetic variants were identified. Comparison of the two DNA sources from each individual showed an average concordance rate of 95.69%. By lowering the variant calling parameters, the concordance rate between the paired samples increased to 99.82%. Sanger sequencing of a subset of the remaining discordant variants did confirm the presence of de novo mutations arising in LCLs.
Conclusions: By varying software stringency parameters, we identified 99% concordance between DNA sequences derived from the two different sources from the same donors. These results suggest that LCLs are an appropriate representation of the genetic material of the donor and suggest that EBV transformation can result in low-level generation of de novo mutations. Therefore, use of PBMC or early passage EBV-transformed cells is recommended. These findings have broad-reaching implications, as there are thousands of LCLs in public biorepositories and individual laboratories.
Figures


References
-
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. - DOI - PMC - PubMed
-
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurrò MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G. ITALSGEN Consortium. Galassi G, Scholz SW, Taylor JP, Restagno G, Chiò A, Traynor BJ. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. - DOI - PMC - PubMed
-
- Anastasio N, Ben-Omran T, Teebi A, Ha KC, Lalonde E, Ali R, Almureikhi M, Der Kaloustian VM, Liu J, Rosenblatt DS, Majewski J, Jerome-Majewska LA. Mutations in SCARF2 are responsible for Van Den Ende-Gupta syndrome. Am J Hum Genet. 2010;87:553–559. doi: 10.1016/j.ajhg.2010.09.005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

