Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania
- PMID: 21944045
- PMCID: PMC3188841
- DOI: 10.1016/j.ajhg.2011.09.005
Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania
Abstract
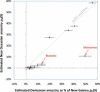
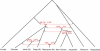
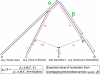
It has recently been shown that ancestors of New Guineans and Bougainville Islanders have inherited a proportion of their ancestry from Denisovans, an archaic hominin group from Siberia. However, only a sparse sampling of populations from Southeast Asia and Oceania were analyzed. Here, we quantify Denisova admixture in 33 additional populations from Asia and Oceania. Aboriginal Australians, Near Oceanians, Polynesians, Fijians, east Indonesians, and Mamanwa (a "Negrito" group from the Philippines) have all inherited genetic material from Denisovans, but mainland East Asians, western Indonesians, Jehai (a Negrito group from Malaysia), and Onge (a Negrito group from the Andaman Islands) have not. These results indicate that Denisova gene flow occurred into the common ancestors of New Guineans, Australians, and Mamanwa but not into the ancestors of the Jehai and Onge and suggest that relatives of present-day East Asians were not in Southeast Asia when the Denisova gene flow occurred. Our finding that descendants of the earliest inhabitants of Southeast Asia do not all harbor Denisova admixture is inconsistent with a history in which the Denisova interbreeding occurred in mainland Asia and then spread over Southeast Asia, leading to all its earliest modern human inhabitants. Instead, the data can be most parsimoniously explained if the Denisova gene flow occurred in Southeast Asia itself. Thus, archaic Denisovans must have lived over an extraordinarily broad geographic and ecological range, from Siberia to tropical Asia.
Copyright © 2011 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Mellars P. Going east: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science. 2006;313:796–800. - PubMed
-
- Lahr M., Foley R. Multiple dispersals and modern human origins. Evol. Anthropol. 1994;3:48–60.
-
- Macaulay V., Hill C., Achilli A., Rengo C., Clarke D., Meehan W., Blackburn J., Semino O., Scozzari R., Cruciani F. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. - PubMed
-
- Thangaraj K., Chaubey G., Kivisild T., Reddy A.G., Singh V.K., Rasalkar A.A., Singh L. Reconstructing the origin of Andaman Islanders. Science. 2005;308:996. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

