Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways
- PMID: 21946159
- PMCID: PMC3256257
- DOI: 10.1016/j.ymben.2011.09.004
Applications of genetically-encoded biosensors for the construction and control of biosynthetic pathways
Abstract
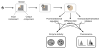
Cells are filled with biosensors, molecular systems that measure the state of the cell and respond by regulating host processes. In much the same way that an engineer would monitor a chemical reactor, the cell uses these sensors to monitor changing intracellular environments and produce consistent behavior despite the variable environment. While natural systems derive a clear benefit from pathway regulation, past research efforts in engineering cellular metabolism have focused on introducing new pathways and removing existing pathway regulation. Synthetic biology is a rapidly growing field that focuses on the development of new tools that support the design, construction, and optimization of biological systems. Recent advances have been made in the design of genetically-encoded biosensors and the application of this class of molecular tools for optimizing and regulating heterologous pathways. Biosensors to cellular metabolites can be taken directly from natural systems, engineered from natural sensors, or constructed entirely in vitro. When linked to reporters, such as antibiotic resistance markers, these metabolite sensors can be used to report on pathway productivity, allowing high-throughput screening for pathway optimization. Future directions will focus on the application of biosensors to introduce feedback control into metabolic pathways, providing dynamic control strategies to increase the efficient use of cellular resources and pathway reliability.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
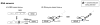


References
-
- Anesiadis N, et al. Dynamic metabolic engineering for increasing bioprocess productivity. Metab Eng. 2008;10:255–66. - PubMed
-
- Ang J, et al. Considerations for using integral feedback control to construct a perfectly adapting synthetic gene network. J Theor Biol. 2010;266:723–38. - PubMed
-
- Atsumi S, et al. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

