Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90
- PMID: 21946277
- PMCID: PMC3265389
- DOI: 10.1038/nchembio.670
Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90
Abstract
Most cancers are characterized by multiple molecular alterations, but identification of the key proteins involved in these signaling pathways is currently beyond reach. We show that the inhibitor PU-H71 preferentially targets tumor-enriched Hsp90 complexes and affinity captures Hsp90-dependent oncogenic client proteins. We have used PU-H71 affinity capture to design a proteomic approach that, when combined with bioinformatic pathway analysis, identifies dysregulated signaling networks and key oncoproteins in chronic myeloid leukemia. The identified interactome overlaps with the well-characterized altered proteome in this cancer, indicating that this method can provide global insights into the biology of individual tumors, including primary patient specimens. In addition, we show that this approach can be used to identify previously uncharacterized oncoproteins and mechanisms, potentially leading to new targeted therapies. We further show that the abundance of the PU-H71-enriched Hsp90 species, which is not dictated by Hsp90 expression alone, is predictive of the cell's sensitivity to Hsp90 inhibition.
Figures


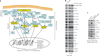
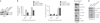
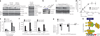
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

