SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities
- PMID: 21947282
- PMCID: PMC3232929
- DOI: 10.1128/MCB.06147-11
SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities
Abstract
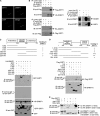
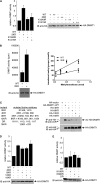
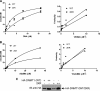
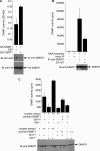
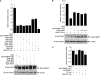
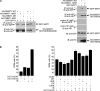
DNA methylation and histone acetylation/deacetylation are distinct biochemical processes that control gene expression. While DNA methylation is a common epigenetic signal that inhibits gene transcription, histone deacetylation similarly represses transcription but can be both an epigenetic and nonepigenetic phenomenon. Here we report that the histone deacetylase SIRT1 regulates the activities of DNMT1, a key enzyme responsible for DNA methylation. In mass spectrometry analysis, 12 new acetylated lysine sites were identified in DNMT1. SIRT1 physically associates with DNMT1 and can deacetylate acetylated DNMT1 in vitro and in vivo. Interestingly, deacetylation of different lysines on DNMT1 has different effects on the functions of DNMT1. For example, deacetylation of Lys1349 and Lys1415 in the catalytic domain of DNMT1 enhances DNMT1's methyltransferase activity, while deacetylation of lysine residues in the GK linker decreases DNMT1's methyltransferase-independent transcriptional repression function. Furthermore, deacetylation of all identified acetylated lysine sites in DNMT1 abrogates its binding to SIRT1 and impairs its capability to regulate cell cycle G(2)/M transition. Finally, inhibition of SIRT1 strengthens the silencing effects of DNMT1 on the expression of tumor suppressor genes ER-α and CDH1 in MDA-MB-231 breast cancer cells. Together, these results suggest that SIRT1-mediated deacetylation of DNMT1 is crucial for DNMT1's multiple effects in gene silencing.
Figures
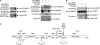


References
-
- Adams R. L., Rinaldi A., Seivwright C. 1991. Microassay for DNA methyltransferase. J. Biochem. Biophys. Methods 22:19–22 - PubMed
-
- Aizawa H., et al. 2004. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303:197–202 - PubMed
-
- Beard C., Li E., Jaenisch R. 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 9:2325–2334 - PubMed
-
- Bender C. M., Pao M. M., Jones P. A. 1998. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 58:95–101 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
