Recurrent deletions and reciprocal duplications of 10q11.21q11.23 including CHAT and SLC18A3 are likely mediated by complex low-copy repeats
- PMID: 21948486
- PMCID: PMC3655525
- DOI: 10.1002/humu.21614
Recurrent deletions and reciprocal duplications of 10q11.21q11.23 including CHAT and SLC18A3 are likely mediated by complex low-copy repeats
Abstract
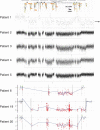
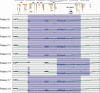
We report 24 unrelated individuals with deletions and 17 additional cases with duplications at 10q11.21q21.1 identified by chromosomal microarray analysis. The rearrangements range in size from 0.3 to 12 Mb. Nineteen of the deletions and eight duplications are flanked by large, directly oriented segmental duplications of >98% sequence identity, suggesting that nonallelic homologous recombination (NAHR) caused these genomic rearrangements. Nine individuals with deletions and five with duplications have additional copy number changes. Detailed clinical evaluation of 20 patients with deletions revealed variable clinical features, with developmental delay (DD) and/or intellectual disability (ID) as the only features common to a majority of individuals. We suggest that some of the other features present in more than one patient with deletion, including hypotonia, sleep apnea, chronic constipation, gastroesophageal and vesicoureteral refluxes, epilepsy, ataxia, dysphagia, nystagmus, and ptosis may result from deletion of the CHAT gene, encoding choline acetyltransferase, and the SLC18A3 gene, mapping in the first intron of CHAT and encoding vesicular acetylcholine transporter. The phenotypic diversity and presence of the deletion in apparently normal carrier parents suggest that subjects carrying 10q11.21q11.23 deletions may exhibit variable phenotypic expressivity and incomplete penetrance influenced by additional genetic and nongenetic modifiers.
© 2011 Wiley Periodicals, Inc.
Figures


References
-
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
-
- Ballif BC, Hornor SA, Jenkins E, Madan-Khetarpal S, Surti U, Jackson KE, Asamoah A, Brock PL, Gowans GC, Conway RL, Graham JM, Jr., Medne L, Zackai EH, Shaikh TH, Geoghegan J, Selzer RR, Eis PS, Bejjani BA, Shaffer LG. Discovery of a previously unrecognized microdeletion syndrome of 16p11.2-p12.2. Nat Genet. 2007;39:1071–1073. - PubMed
-
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, McDonald M, Campbell L, Ming JE, Zackai EH, Bejjani BA, Shaffer LG. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008a;1:8. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

