Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes
- PMID: 21948829
- PMCID: PMC3209000
- DOI: 10.1128/AEM.05439-11
Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes
Abstract
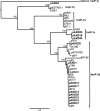
Broad-host-range plasmids are known to spread genes between distinct phylogenetic groups of bacteria. These genes often code for resistances to antibiotics and heavy metals or degradation of pollutants. Although some broad-host-range plasmids have been extensively studied, their evolutionary history and genetic diversity remain largely unknown. The goal of this study was to analyze and compare the genomes of 12 broad-host-range plasmids that were previously isolated from Norwegian soils by exogenous plasmid isolation and that encode mercury resistance. Complete nucleotide sequencing followed by phylogenetic analyses based on the relaxase gene traI showed that all the plasmids belong to one of two subgroups (β and ε) of the well-studied incompatibility group IncP-1. A diverse array of accessory genes was found to be involved in resistance to antimicrobials (streptomycin, spectinomycin, and sulfonamides), degradation of herbicides (2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenoxypropionic acid), and a putative new catabolic pathway. Intramolecular transposition of insertion sequences followed by deletion was found to contribute to the diversity of some of these plasmids. The previous observation that the insertion sites of a Tn501-related element are identical in four IncP-1β plasmids (pJP4, pB10, R906, and R772) was further extended to three more IncP-1β plasmids (pAKD15, pAKD18, and pAKD29). We proposed a hypothesis for the evolution of these Tn501-bearing IncP-1β plasmids that predicts recent diversification followed by worldwide spread. Our study increases the available collection of complete IncP-1 plasmid genome sequences by 50% and will aid future studies to enhance our understanding of the evolution and function of this important plasmid family.
Figures




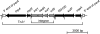
Similar articles
-
The 64 508 bp IncP-1beta antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1beta group.Microbiology (Reading). 2003 Nov;149(Pt 11):3139-3153. doi: 10.1099/mic.0.26570-0. Microbiology (Reading). 2003. PMID: 14600226
-
Diversification of broad host range plasmids correlates with the presence of antibiotic resistance genes.FEMS Microbiol Ecol. 2016 Jan;92(1):fiv151. doi: 10.1093/femsec/fiv151. Epub 2015 Dec 2. FEMS Microbiol Ecol. 2016. PMID: 26635412 Free PMC article.
-
The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1beta group without any accessory genes.Microbiology (Reading). 2004 Nov;150(Pt 11):3591-3599. doi: 10.1099/mic.0.27304-0. Microbiology (Reading). 2004. PMID: 15528648
-
Comparison of the organisation of the genomes of phenotypically diverse plasmids of incompatibility group P: members of the IncP beta sub-group are closely related.Mol Gen Genet. 1987 Mar;206(3):419-27. doi: 10.1007/BF00428881. Mol Gen Genet. 1987. PMID: 3035342
-
The evolution of IncP catabolic plasmids.Curr Opin Biotechnol. 2005 Jun;16(3):291-8. doi: 10.1016/j.copbio.2005.04.002. Curr Opin Biotechnol. 2005. PMID: 15961030 Review.
Cited by
-
Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy.Front Microbiol. 2015 Mar 31;6:242. doi: 10.3389/fmicb.2015.00242. eCollection 2015. Front Microbiol. 2015. PMID: 25873913 Free PMC article. Review.
-
Widespread dissemination of class 1 integron components in soils and related ecosystems as revealed by cultivation-independent analysis.Front Microbiol. 2014 Jan 17;4:420. doi: 10.3389/fmicb.2013.00420. eCollection 2013. Front Microbiol. 2014. PMID: 24478761 Free PMC article.
-
Plasmid-Encoded Traits Vary across Environments.mBio. 2023 Feb 28;14(1):e0319122. doi: 10.1128/mbio.03191-22. Epub 2023 Jan 11. mBio. 2023. PMID: 36629415 Free PMC article.
-
Non-antibiotic pharmaceuticals promote conjugative plasmid transfer at a community-wide level.Microbiome. 2022 Aug 12;10(1):124. doi: 10.1186/s40168-022-01314-y. Microbiome. 2022. PMID: 35953866 Free PMC article.
-
Shifts in abundance and diversity of mobile genetic elements after the introduction of diverse pesticides into an on-farm biopurification system over the course of a year.Appl Environ Microbiol. 2014 Jul;80(13):4012-20. doi: 10.1128/AEM.04016-13. Epub 2014 Apr 25. Appl Environ Microbiol. 2014. PMID: 24771027 Free PMC article.
References
-
- Bahl M. I., Hansen L. H., Goesmann A., Sørensen S. J. 2007. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established alpha, beta and delta sub-groups. Plasmid 58:31–43 - PubMed
-
- Baker-Austin C., Wright M. S., Stepanauskas R., McArthur J. V. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176–182 - PubMed
-
- Reference deleted.
-
- Chattoraj D. K. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467–476 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

