Functional genomic screen and network analysis reveal novel modifiers of tauopathy dissociated from tau phosphorylation
- PMID: 21949350
- PMCID: PMC3221533
- DOI: 10.1093/hmg/ddr432
Functional genomic screen and network analysis reveal novel modifiers of tauopathy dissociated from tau phosphorylation
Abstract
A functional genetic screen using loss-of-function and gain-of-function alleles was performed to identify modifiers of tau-induced neurotoxicity using the 2N/4R (full-length) isoform of wild-type human tau expressed in the fly retina. We previously reported eye pigment mutations, which create dysfunctional lysosomes, as potent modifiers; here, we report 37 additional genes identified from ∼1900 genes screened, including the kinases shaggy/GSK-3beta, par-1/MARK, CamKI and Mekk1. Tau acts synergistically with Mekk1 and p38 to down-regulate extracellular regulated kinase activity, with a corresponding decrease in AT8 immunoreactivity (pS202/T205), suggesting that tau can participate in signaling pathways to regulate its own kinases. Modifiers showed poor correlation with tau phosphorylation (using the AT8, 12E8 and AT270 epitopes); moreover, tested suppressors of wild-type tau were equally effective in suppressing toxicity of a phosphorylation-resistant S11A tau construct, demonstrating that changes in tau phosphorylation state are not required to suppress or enhance its toxicity. Genes related to autophagy, the cell cycle, RNA-associated proteins and chromatin-binding proteins constitute a large percentage of identified modifiers. Other functional categories identified include mitochondrial proteins, lipid trafficking, Golgi proteins, kinesins and dynein and the Hsp70/Hsp90-organizing protein (Hop). Network analysis uncovered several other genes highly associated with the functional modifiers, including genes related to the PI3K, Notch, BMP/TGF-β and Hedgehog pathways, and nuclear trafficking. Activity of GSK-3β is strongly upregulated due to TDP-43 expression, and reduced GSK-3β dosage is also a common suppressor of Aβ42 and TDP-43 toxicity. These findings suggest therapeutic targets other than mitigation of tau phosphorylation.
Figures







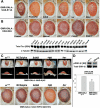
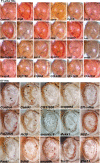
Comment in
-
The downward spiral of tau and autolysosomes: a new hypothesis in neurodegeneration.Autophagy. 2012 Jul 1;8(7):1144-5. doi: 10.4161/auto.20515. Epub 2012 May 31. Autophagy. 2012. PMID: 22635052 Free PMC article.
Similar articles
-
Dissociation of tau toxicity and phosphorylation: role of GSK-3beta, MARK and Cdk5 in a Drosophila model.Hum Mol Genet. 2009 Jan 1;18(1):164-77. doi: 10.1093/hmg/ddn326. Epub 2008 Oct 17. Hum Mol Genet. 2009. PMID: 18930955 Free PMC article.
-
Nav1.7-Ca2+ influx-induced increased phosphorylations of extracellular signal-regulated kinase (ERK) and p38 attenuate tau phosphorylation via glycogen synthase kinase-3beta: priming of Nav1.7 gating by ERK and p38.Eur J Pharmacol. 2010 Aug 25;640(1-3):20-8. doi: 10.1016/j.ejphar.2010.04.048. Epub 2010 May 12. Eur J Pharmacol. 2010. PMID: 20470771
-
Inhibition of glycogen synthase kinase-3β by Angelica sinensis extract decreases β-amyloid-induced neurotoxicity and tau phosphorylation in cultured cortical neurons.J Neurosci Res. 2011 Mar;89(3):437-47. doi: 10.1002/jnr.22563. Epub 2010 Dec 17. J Neurosci Res. 2011. PMID: 21259330
-
Cellular and molecular modifier pathways in tauopathies: the big picture from screening invertebrate models.J Neurochem. 2016 Apr;137(1):12-25. doi: 10.1111/jnc.13532. Epub 2016 Feb 11. J Neurochem. 2016. PMID: 26756400 Review.
-
Current advances on different kinases involved in tau phosphorylation, and implications in Alzheimer's disease and tauopathies.Curr Alzheimer Res. 2005 Jan;2(1):3-18. doi: 10.2174/1567205052772713. Curr Alzheimer Res. 2005. PMID: 15977985 Review.
Cited by
-
With or without You: Co-Chaperones Mediate Health and Disease by Modifying Chaperone Function and Protein Triage.Cells. 2021 Nov 11;10(11):3121. doi: 10.3390/cells10113121. Cells. 2021. PMID: 34831344 Free PMC article. Review.
-
The synaptic maintenance problem: membrane recycling, Ca2+ homeostasis and late onset degeneration.Mol Neurodegener. 2013 Jul 8;8:23. doi: 10.1186/1750-1326-8-23. Mol Neurodegener. 2013. PMID: 23829673 Free PMC article. Review.
-
Quantitative Assessment of Eye Phenotypes for Functional Genetic Studies Using Drosophila melanogaster.G3 (Bethesda). 2016 May 3;6(5):1427-37. doi: 10.1534/g3.116.027060. G3 (Bethesda). 2016. PMID: 26994292 Free PMC article.
-
Autophagy and Human Neurodegenerative Diseases-A Fly's Perspective.Int J Mol Sci. 2017 Jul 23;18(7):1596. doi: 10.3390/ijms18071596. Int J Mol Sci. 2017. PMID: 28737703 Free PMC article. Review.
-
Early Steps of Mammary Stem Cell Transformation by Exogenous Signals; Effects of Bisphenol Endocrine Disrupting Chemicals and Bone Morphogenetic Proteins.Cancers (Basel). 2019 Sep 12;11(9):1351. doi: 10.3390/cancers11091351. Cancers (Basel). 2019. PMID: 31547326 Free PMC article. Review.
References
-
- Iqbal K., Alonso A.D.C., Chen S., Chohan M.O., El-Akkad E., Gong C.-X., Khatoon S., Li B., Liu F., Rahman A., et al. Tau pathology in Alzheimer disease and other tauopathies. Biochim. Biophys. Acta. 2005;1739:198–210. - PubMed
-
- Galpern W.R., Lang A.E. Interface between tauopathies and synucleinopathies: a tale of two proteins. Ann. Neurol. 2006;59:449–458. doi:10.1002/ana.20819. - DOI - PubMed
-
- Hutton M., Lendon C.L., Rizzu P., Baker M., Froelich S., Houlden H., Pickering-Brown S., Chakraverty S., Isaacs A., Grover A., et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi:10.1038/31508. - DOI - PubMed
-
- Poorkaj P., Bird T.D., Wijsman E., Nemens E., Garruto R.M., Anderson L., Andreadis A., Wiederholt W.C., Raskind M., Schellenberg G.D. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol. 1998;43:815–825. doi:10.1002/ana.410430617. - DOI - PubMed
-
- Binder L.I., Frankfurter A., Rebhun L.I. The distribution of tau in the mammalian central nervous system. J. Cell Biol. 1985;101:1371–1378. doi:10.1083/jcb.101.4.1371. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

