UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity
- PMID: 21949367
- PMCID: PMC3219150
- DOI: 10.1073/pnas.1113170108
UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity
Abstract
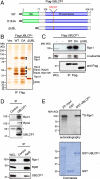
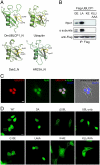
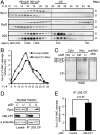
Protein degradation by the 26S proteasome is a fundamental process involved in a broad range of cellular activities, yet how proteasome activity is regulated remains poorly understood. We report here that ubiquitin-like domain-containing C-terminal domain phosphatase 1 (UBLCP1) is a 26S proteasome phosphatase that regulates nuclear proteasome activity. UBLCP1 directly interacts with the proteasome via its UBL domain and is exclusively localized in the nucleus. UBLCP1 dephosphorylates the 26S proteasome and inhibits proteasome activity in vitro. Knockdown of UBLCP1 in cells promotes 26S proteasome assembly and selectively enhances nuclear proteasome activity. Our results describe the first identified proteasome-specific phosphatase and uncover a unique mechanism for phosphoregulation of the proteasome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Keeping proteasomes under control--a role for phosphorylation in the nucleus.Proc Natl Acad Sci U S A. 2011 Nov 15;108(46):18573-4. doi: 10.1073/pnas.1115315108. Epub 2011 Nov 7. Proc Natl Acad Sci U S A. 2011. PMID: 22065770 Free PMC article. No abstract available.
References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
-
- Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10(Suppl):S2–S9. - PubMed
-
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. - PubMed
-
- Bingol B, Sheng M. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

