A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis
- PMID: 21949371
- PMCID: PMC3203754
- DOI: 10.1073/pnas.1100132108
A whole-genome RNAi screen identifies an 8q22 gene cluster that inhibits death receptor-mediated apoptosis
Abstract
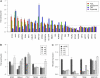
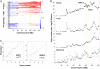
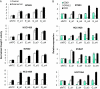
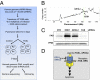
Deregulation of apoptosis is a common occurrence in cancer, for which emerging oncology therapeutic agents designed to engage this pathway are undergoing clinical trials. With the aim of uncovering strategies to activate apoptosis in cancer cells, we used a pooled shRNA screen to interrogate death receptor signaling. This screening approach identified 16 genes that modulate the sensitivity to ligand induced apoptosis, with several genes exhibiting frequent overexpression and/or copy number gain in cancer. Interestingly, two of the top hits, EDD1 and GRHL2, are found 50 kb apart on chromosome 8q22, a region that is frequently amplified in many cancers. By using a series of silencing and overexpression studies, we show that EDD1 and GRHL2 suppress death-receptor expression, and that EDD1 expression is elevated in breast, pancreas, and lung cancer cell lines resistant to death receptor-mediated apoptosis. Supporting the relevance of EDD1 and GRHL2 as therapeutic candidates to engage apoptosis in cancer cells, silencing the expression of either gene sensitizes 8q22-amplified breast cancer cell lines to death receptor induced apoptosis. Our findings highlight a mechanism by which cancer cells may evade apoptosis, and therefore provide insight in the search for new targets and functional biomarkers for this pathway.
Conflict of interest statement
Conflict of interest statement: All the authors are employees of Genentech.
Figures



References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Ashkenazi A. Directing cancer cells to self-destruct with pro-apoptotic receptor agonists. Nat Rev Drug Discov. 2008;7:1001–1012. - PubMed
-
- Ashkenazi A, Dixit VM. Death receptors: Signaling and modulation. Science. 1998;281:1305–1308. - PubMed
-
- Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

