Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development
- PMID: 21949639
- PMCID: PMC3176755
- DOI: 10.1371/journal.pbio.1001155
Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development
Abstract
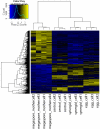
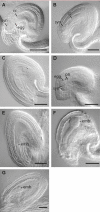
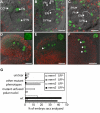
Germ line specification is a crucial step in the life cycle of all organisms. For sexual plant reproduction, the megaspore mother cell (MMC) is of crucial importance: it marks the first cell of the plant "germline" lineage that gets committed to undergo meiosis. One of the meiotic products, the functional megaspore, subsequently gives rise to the haploid, multicellular female gametophyte that harbours the female gametes. The MMC is formed by selection and differentiation of a single somatic, sub-epidermal cell in the ovule. The transcriptional network underlying MMC specification and differentiation is largely unknown. We provide the first transcriptome analysis of an MMC using the model plant Arabidopsis thaliana with a combination of laser-assisted microdissection and microarray hybridizations. Statistical analyses identified an over-representation of translational regulation control pathways and a significant enrichment of DEAD/DEAH-box helicases in the MMC transcriptome, paralleling important features of the animal germline. Analysis of two independent T-DNA insertion lines suggests an important role of an enriched helicase, MNEME (MEM), in MMC differentiation and the restriction of the germline fate to only one cell per ovule primordium. In heterozygous mem mutants, additional enlarged MMC-like cells, which sometimes initiate female gametophyte development, were observed at higher frequencies than in the wild type. This closely resembles the phenotype of mutants affected in the small RNA and DNA-methylation pathways important for epigenetic regulation. Importantly, the mem phenotype shows features of apospory, as female gametophytes initiate from two non-sister cells in these mutants. Moreover, in mem gametophytic nuclei, both higher order chromatin structure and the distribution of LIKE HETEROCHROMATIN PROTEIN1 were affected, indicating epigenetic perturbations. In summary, the MMC transcriptome sets the stage for future functional characterization as illustrated by the identification of MEM, a novel gene involved in the restriction of germline fate.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Grossniklaus U. Plant germline development: a tale of cross-talk, signaling, and cellular interactions. Sex Plant Reprod. 2011;24:91–95. - PubMed
-
- Grossniklaus U, Schneitz K. The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol. 1998;9:227–238. - PubMed
-
- Koltunow A. M, Grossniklaus U. Apomixis: a developmental perspective. Annu Rev Plant Biol. 2003;54:547–574. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

