Inactivation of Pmel alters melanosome shape but has only a subtle effect on visible pigmentation
- PMID: 21949658
- PMCID: PMC3174228
- DOI: 10.1371/journal.pgen.1002285
Inactivation of Pmel alters melanosome shape but has only a subtle effect on visible pigmentation
Abstract
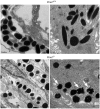
PMEL is an amyloidogenic protein that appears to be exclusively expressed in pigment cells and forms intralumenal fibrils within early stage melanosomes upon which eumelanins deposit in later stages. PMEL is well conserved among vertebrates, and allelic variants in several species are associated with reduced levels of eumelanin in epidermal tissues. However, in most of these cases it is not clear whether the allelic variants reflect gain-of-function or loss-of-function, and no complete PMEL loss-of-function has been reported in a mammal. Here, we have created a mouse line in which the Pmel gene has been inactivated (Pmel⁻/⁻). These mice are fully viable, fertile, and display no obvious developmental defects. Melanosomes within Pmel⁻/⁻ melanocytes are spherical in contrast to the oblong shape present in wild-type animals. This feature was documented in primary cultures of skin-derived melanocytes as well as in retinal pigment epithelium cells and in uveal melanocytes. Inactivation of Pmel has only a mild effect on the coat color phenotype in four different genetic backgrounds, with the clearest effect in mice also carrying the brown/Tyrp1 mutation. This phenotype, which is similar to that observed with the spontaneous silver mutation in mice, strongly suggests that other previously described alleles in vertebrates with more striking effects on pigmentation are dominant-negative mutations. Despite a mild effect on visible pigmentation, inactivation of Pmel led to a substantial reduction in eumelanin content in hair, which demonstrates that PMEL has a critical role for maintaining efficient epidermal pigmentation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Ito S, Wakamatsu K. Chemistry of mixed melanogenesis--pivotal roles of dopaquinone. Photochem Photobiol. 2008;84:582–592. - PubMed
-
- Baxter LL, Pavan WJ. Pmel17 expression is Mitf-dependent and reveals cranial melanoblast migration during murine development. Gene Expr Patterns. 2003;3:703–707. - PubMed
-
- Chakraborty AK, Platt JT, Kim KK, Kwon BS, Bennett DC, et al. Polymerization of 5,6-dihydroxyindole-2-carboxylic acid to melanin by the pmel 17/silver locus protein. Eur J Biochem. 1996;236:180–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

