Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia)
- PMID: 21949820
- PMCID: PMC3173496
- DOI: 10.1371/journal.pone.0024984
Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia)
Abstract
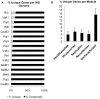
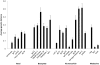
Genome evolution of bacteria is usually influenced by ecology, such that bacteria with a free-living stage have large genomes and high rates of horizontal gene transfer, while obligate intracellular bacteria have small genomes with typically low amounts of gene exchange. However, recent studies indicate that obligate intracellular species that host-switch frequently harbor agents of horizontal transfer such as mobile elements. For example, the temperate double-stranded DNA bacteriophage WO in Wolbachia persistently transfers between bacterial coinfections in the same host. Here we show that despite the phage's rampant mobility between coinfections, the prophage's genome displays features of constraint related to its intracellular niche. First, there is always at least one intact prophage WO and usually several degenerate, independently-acquired WO prophages in each Wolbachia genome. Second, while the prophage genomes are modular in composition with genes of similar function grouping together, the modules are generally not interchangeable with other unrelated phages and thus do not evolve by the Modular Theory. Third, there is an unusual core genome that strictly consists of head and baseplate genes; other gene modules are frequently deleted. Fourth, the prophage recombinases are diverse and there is no conserved integration sequence. Finally, the molecular evolutionary forces acting on prophage WO are point mutation, intragenic recombination, deletion, and purifying selection. Taken together, these analyses indicate that while lateral transfer of phage WO is pervasive between Wolbachia with occasional new gene uptake, constraints of the intracellular niche obstruct extensive mixture between WO and the global phage population. Although the Modular Theory has long been considered the paradigm of temperate bacteriophage evolution in free-living bacteria, it appears irrelevant in phages of obligate intracellular bacteria.
Conflict of interest statement
Figures



Similar articles
-
Lateral phage transfer in obligate intracellular bacteria (wolbachia): verification from natural populations.Mol Biol Evol. 2010 Mar;27(3):501-5. doi: 10.1093/molbev/msp275. Epub 2009 Nov 11. Mol Biol Evol. 2010. PMID: 19906794 Free PMC article.
-
Muramidase, nuclease, or hypothetical protein genes intervene between paired genes encoding DNA packaging terminase and portal proteins in Wolbachia phages and prophages.Virus Genes. 2022 Aug;58(4):327-349. doi: 10.1007/s11262-022-01907-7. Epub 2022 May 10. Virus Genes. 2022. PMID: 35538383
-
Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture.Genome Biol Evol. 2011;3:209-18. doi: 10.1093/gbe/evr007. Epub 2011 Feb 2. Genome Biol Evol. 2011. PMID: 21292630 Free PMC article.
-
Phage WO of Wolbachia: lambda of the endosymbiont world.Trends Microbiol. 2010 Apr;18(4):173-81. doi: 10.1016/j.tim.2009.12.011. Epub 2010 Jan 18. Trends Microbiol. 2010. PMID: 20083406 Free PMC article. Review.
-
Phage as agents of lateral gene transfer.Curr Opin Microbiol. 2003 Aug;6(4):417-24. doi: 10.1016/s1369-5274(03)00086-9. Curr Opin Microbiol. 2003. PMID: 12941415 Review.
Cited by
-
Proteomic profiling of a robust Wolbachia infection in an Aedes albopictus mosquito cell line.Mol Microbiol. 2014 Nov;94(3):537-56. doi: 10.1111/mmi.12768. Epub 2014 Sep 22. Mol Microbiol. 2014. PMID: 25155417 Free PMC article.
-
The complexity of virus systems: the case of endosymbionts.Curr Opin Microbiol. 2012 Aug;15(4):546-52. doi: 10.1016/j.mib.2012.04.010. Epub 2012 May 19. Curr Opin Microbiol. 2012. PMID: 22609369 Free PMC article.
-
Complete De Novo Assembly of Wolbachia Endosymbiont of Frankliniella intonsa.Int J Mol Sci. 2023 Aug 26;24(17):13245. doi: 10.3390/ijms241713245. Int J Mol Sci. 2023. PMID: 37686049 Free PMC article.
-
The impact of artificial selection for Wolbachia-mediated dengue virus blocking on phage WO.PLoS Negl Trop Dis. 2021 Jul 27;15(7):e0009637. doi: 10.1371/journal.pntd.0009637. eCollection 2021 Jul. PLoS Negl Trop Dis. 2021. PMID: 34314434 Free PMC article.
-
High Levels of Multiple Phage WO Infections and Its Evolutionary Dynamics Associated With Wolbachia-Infected Butterflies.Front Microbiol. 2022 Apr 21;13:865227. doi: 10.3389/fmicb.2022.865227. eCollection 2022. Front Microbiol. 2022. PMID: 35531293 Free PMC article.
References
-
- Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–424. - PubMed
-
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. - PubMed
-
- Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, et al. Explaining microbial population genomics through phage predation. Nature Reviews Microbiology. 2009;7:828–836. - PubMed
-
- Brussow H, Hendrix RW. Phage genomics: small is beautiful. Cell. 2002;108:13–16. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

