Functional consequences of the creation of an Asp-His-Fe triad in a 3/3 globin
- PMID: 21950839
- PMCID: PMC4007314
- DOI: 10.1021/bi201368u
Functional consequences of the creation of an Asp-His-Fe triad in a 3/3 globin
Abstract
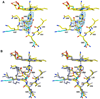
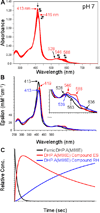
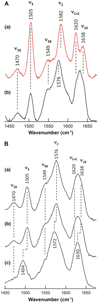
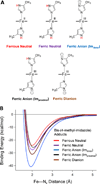
The proximal side of dehaloperoxidase-hemoglobin A (DHP A) from Amphitrite ornata has been modified via site-directed mutagenesis of methionine 86 into aspartate (M86D) to introduce an Asp-His-Fe triad charge relay. X-ray crystallographic structure determination of the metcyano forms of M86D [Protein Data Bank (PDB) entry 3MYN ] and M86E (PDB entry 3MYM ) mutants reveal the structural origins of a stable catalytic triad in DHP A. A decrease in the rate of H(2)O(2) activation as well as a lowered reduction potential versus that of the wild-type enzyme was observed in M86D. One possible explanation for the significantly lower activity is an increased affinity for the distal histidine in binding to the heme Fe to form a bis-histidine adduct. Resonance Raman spectroscopy demonstrates a pH-dependent ligation by the distal histidine in M86D, which is indicative of an increased trans effect. At pH 5.0, the heme Fe is five-coordinate, and this structure resembles the wild-type DHP A resting state. However, at pH 7.0, the distal histidine appears to form a six-coordinate ferric bis-histidine (hemichrome) adduct. These observations can be explained by the effect of the increased positive charge on the heme Fe on the formation of a six-coordinate low-spin adduct, which inhibits the ligation and activation of H(2)O(2) as required for peroxidase activity. The results suggest that the proximal charge relay in peroxidases regulate the redox potential of the heme Fe but that the trans effect is a carefully balanced property that can both activate H(2)O(2) and attract ligation by the distal histidine. To understand the balance of forces that modulate peroxidase reactivity, we studied three M86 mutants, M86A, M86D, and M86E, by spectroelectrochemistry and nuclear magnetic resonance spectroscopy of (13)C- and (15)N-labeled cyanide adducts as probes of the redox potential and of the trans effect in the heme Fe, both of which can be correlated with the proximity of negative charge to the N(δ) hydrogen of the proximal histidine, consistent with an Asp-His-Fe charge relay observed in heme peroxidases.
Figures






References
-
- Chen YP, Woodin SA, Lincoln DE, Lovell CR. An unusual dehalogenating peroxidase from the marine terebellid polychaete amphitrite ornata. J. Biol. Chem. 1996;271:4609–4612. - PubMed
-
- Lebioda L, LaCount MW, Zhang E, Chen YP, Han KP, Whitton MM, Lincoln DE, Woodin SA. An enzymatic globin from a marine worm. Nature. 1999;401:445. - PubMed
-
- LaCount MW, Zhang EL, Chen YP, Han KP, Whitton MM, Lincoln DE, Woodin SA, Lebioda L. The crystal structure and amino acid sequence of dehaloperoxidase from amphitrite ornata indicate common ancestry with globins. J. Biol. Chem. 2000;275:18712–18716. - PubMed
-
- Belyea J, Gilvey LB, Davis MF, Godek M, Sit TL, Lommel SA, Franzen S. Enzyme function of the globin dehaloperoxidase from amphitrite ornata is activated by substrate binding. Biochemistry. 2005;44:15637–15644. - PubMed
-
- Bashford D, Chothia C, Lesk AM. Determinants of a protein fold - unique features of the globin amino-acid-sequences. J. Mol. Biol. 1987;196:199–216. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

