The use of residual dipolar coupling in studying proteins by NMR
- PMID: 21952837
- PMCID: PMC4153736
- DOI: 10.1007/128_2011_215
The use of residual dipolar coupling in studying proteins by NMR
Abstract
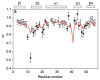
The development of residual dipolar coupling (RDC) in protein NMR spectroscopy, over a decade ago, has become a useful and almost routine tool for accurate protein solution structure determination. RDCs provide orientation information of magnetic dipole-dipole interaction vectors within a common reference frame. Its measurement requires a nonisotropic orientation, through a direct or indirect magnetic field alignment, of the protein in solution. There has been recent progress in the developments of alignment methods to allow the measurement of RDC and of methods to analyze the resulting data. In this chapter we briefly go through the mathematical expressions for the RDC and common descriptions of the alignment tensor, which may be represented using either Saupe order or the principal order matrix. Then we review the latest developments in alignment media. In particular we looked at the lipid-compatible media that allow the measurement of RDCs for membrane proteins. Other methods including conservative surface residue mutation have been invented to obtain up to five orthogonal alignment tensors that provide a potential for de novo structure and dynamics study using RDCs exclusively. We then discuss approximations assumed in RDC interpretations and different views on dynamics uncovered from the RDC method. In addition to routine usage of RDCs in refining a single structure, novel applications such as ensemble refinement against RDCs have been implemented to represent protein structure and dynamics in solution. The RDC application also extends to the study of protein-substrate interaction as well as to solving quaternary structure of oligomer in equilibrium with a monomer, opening an avenue for RDCs in high-order protein structure determination.
Figures
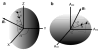

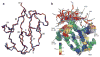

References
-
- Wuthrich K. Determination of 3-dimensional protein structures in solution by nuclear-magnetic-resonance – an overview. Methods Enzymol. 1989;177:125–131. - PubMed
-
- Bax A. Two-dimensional NMR and protein-structure. Annu Rev Biochem. 1989;58:223–256. - PubMed
-
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear-magnetic-resonance of proteins. Nucl Magn Reson Pt C. 1994;239:349–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

