Regulation of IL-2 gene expression by Siva and FOXP3 in human T cells
- PMID: 21955384
- PMCID: PMC3208582
- DOI: 10.1186/1471-2172-12-54
Regulation of IL-2 gene expression by Siva and FOXP3 in human T cells
Abstract
Background: Severe autoinflammatory diseases are associated with mutations in the Foxp3 locus in both mice and humans. Foxp3 is required for the development, function, and maintenance of regulatory T cells (Tregs), a subset of CD4 cells that suppress T cell activation and inflammatory processes. Siva is a pro-apoptotic gene that is expressed across a range of tissues, including CD4 T cells. Siva interacts with three tumor necrosis factor receptor (TNFR) family members that are constitutively expressed on Treg cells: CD27, GITR, and OX40.
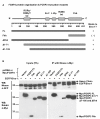
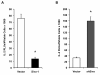
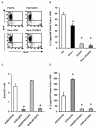
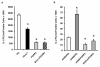
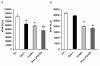
Results: Here we report a biophysical interaction between FOXP3 and Siva. We mapped the interaction domains to Siva's C-terminus and to a central region of FOXP3. We showed that Siva repressed IL-2 induction by suppressing IL-2 promoter activity during T cell activation. Siva-1's repressive effect on IL-2 gene expression appears to be mediated by inhibition of NFkappaB, whereas FOXP3 repressed both NFkappaB and NFAT activity.
Conclusions: In summary, our data suggest that both FOXP3 and Siva function as negative regulators of IL-2 gene expression in Treg cells, via suppression of NFAT by FOXP3 and of NFkappaB by both FOXP3 and Siva. Our work contributes evidence for Siva's role as a T cell signalling mediator in addition to its known pro-apoptotic function. Though further investigations are needed, evidence for the biophysical interaction between FOXP3 and Siva invites the possibility that Siva may be important for proper Treg cell function.
Figures


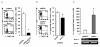
Similar articles
-
Inhibition of FOXP3/NFAT Interaction Enhances T Cell Function after TCR Stimulation.J Immunol. 2015 Oct 1;195(7):3180-9. doi: 10.4049/jimmunol.1402997. Epub 2015 Aug 31. J Immunol. 2015. PMID: 26324768
-
Constitutive nuclear localization of NFAT in Foxp3+ regulatory T cells independent of calcineurin activity.J Immunol. 2012 May 1;188(9):4268-77. doi: 10.4049/jimmunol.1102376. Epub 2012 Apr 4. J Immunol. 2012. PMID: 22490438
-
Gene expression in the Gitr locus is regulated by NF-κB and Foxp3 through an enhancer.J Immunol. 2014 Apr 15;192(8):3915-24. doi: 10.4049/jimmunol.1302174. Epub 2014 Mar 14. J Immunol. 2014. PMID: 24634496 Free PMC article.
-
Control of regulatory T-cell differentiation and function by T-cell receptor signalling and Foxp3 transcription factor complexes.Immunology. 2020 May;160(1):24-37. doi: 10.1111/imm.13178. Epub 2020 Mar 9. Immunology. 2020. PMID: 32022254 Free PMC article. Review.
-
FOXP3 and NFAT: partners in tolerance.Cell. 2006 Jul 28;126(2):253-6. doi: 10.1016/j.cell.2006.07.005. Cell. 2006. PMID: 16873058 Review.
Cited by
-
Molecular Mechanisms Controlling Foxp3 Expression in Health and Autoimmunity: From Epigenetic to Post-translational Regulation.Front Immunol. 2020 Feb 3;10:3136. doi: 10.3389/fimmu.2019.03136. eCollection 2019. Front Immunol. 2020. PMID: 32117202 Free PMC article. Review.
-
An Engineered Probiotic Produces a Type III Interferon IFNL1 and Reduces Inflammations in in vitro Inflammatory Bowel Disease Models.ACS Biomater Sci Eng. 2023 Sep 11;9(9):5123-5135. doi: 10.1021/acsbiomaterials.2c00202. Epub 2022 Nov 18. ACS Biomater Sci Eng. 2023. PMID: 36399014 Free PMC article.
-
SIVA-1 interaction with PCBP1 serves as a predictive biomarker for cisplatin sensitivity in gastric cancer and its inhibitory effect on tumor growth in vivo.J Cancer. 2024 Jun 11;15(13):4301-4312. doi: 10.7150/jca.92963. eCollection 2024. J Cancer. 2024. PMID: 38947376 Free PMC article.
-
Splicing factor SF2/ASF rescues IL-2 production in T cells from systemic lupus erythematosus patients by activating IL-2 transcription.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1845-50. doi: 10.1073/pnas.1214207110. Epub 2013 Jan 14. Proc Natl Acad Sci U S A. 2013. PMID: 23319613 Free PMC article.
-
The Importance of the Transcription Factor Foxp3 in the Development of Primary Immunodeficiencies.J Clin Med. 2022 Feb 11;11(4):947. doi: 10.3390/jcm11040947. J Clin Med. 2022. PMID: 35207219 Free PMC article. Review.
References
-
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L. et al.X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. - DOI - PubMed
-
- Kobayashi I, Shiari R, Yamada M, Kawamura N, Okano M, Yara A, Iguchi A, Ishikawa N, Ariga T, Sakiyama Y. et al.Novel mutations of FOXP3 in two Japanese patients with immune dysregulation, polyendocrinopathy, enteropathy, X linked syndrome (IPEX) J Med Genet. 2001;38(12):874–876. doi: 10.1136/jmg.38.12.874. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

