First somatic mutation of E2F1 in a critical DNA binding residue discovered in well-differentiated papillary mesothelioma of the peritoneum
- PMID: 21955916
- PMCID: PMC3308059
- DOI: 10.1186/gb-2011-12-9-r96
First somatic mutation of E2F1 in a critical DNA binding residue discovered in well-differentiated papillary mesothelioma of the peritoneum
Abstract
Background: Well differentiated papillary mesothelioma of the peritoneum (WDPMP) is a rare variant of epithelial mesothelioma of low malignancy potential, usually found in women with no history of asbestos exposure. In this study, we perform the first exome sequencing of WDPMP.
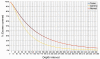
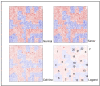
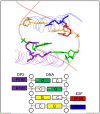
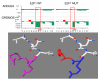
Results: WDPMP exome sequencing reveals the first somatic mutation of E2F1, R166H, to be identified in human cancer. The location is in the evolutionarily conserved DNA binding domain and computationally predicted to be mutated in the critical contact point between E2F1 and its DNA target. We show that the R166H mutation abrogates E2F1's DNA binding ability and is associated with reduced activation of E2F1 downstream target genes. Mutant E2F1 proteins are also observed in higher quantities when compared with wild-type E2F1 protein levels and the mutant protein's resistance to degradation was found to be the cause of its accumulation within mutant over-expressing cells. Cells over-expressing wild-type E2F1 show decreased proliferation compared to mutant over-expressing cells, but cell proliferation rates of mutant over-expressing cells were comparable to cells over-expressing the empty vector.
Conclusions: The R166H mutation in E2F1 is shown to have a deleterious effect on its DNA binding ability as well as increasing its stability and subsequent accumulation in R166H mutant cells. Based on the results, two compatible theories can be formed: R166H mutation appears to allow for protein over-expression while minimizing the apoptotic consequence and the R166H mutation may behave similarly to SV40 large T antigen, inhibiting tumor suppressive functions of retinoblastoma protein 1.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

