Mechanisms of deadenylation-dependent decay
- PMID: 21957004
- PMCID: PMC3624885
- DOI: 10.1002/wrna.40
Mechanisms of deadenylation-dependent decay
Abstract
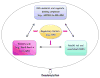
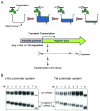
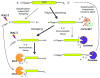
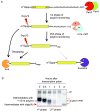
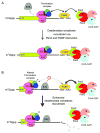
Degradation of messenger RNAs (mRNAs) plays an essential role in modulation of gene expression and in quality control of mRNA biogenesis. Nearly all major mRNA decay pathways characterized thus far in eukaryotes are initiated by deadenylation, i.e., shortening of the mRNA 3(') poly(A) tail. Deadenylation is often a rate-limiting step for mRNA degradation and translational silencing, making it an important control point for both processes. In this review, we discuss the fundamental principles that govern mRNA deadenylation in eukaryotes. We use several major mRNA decay pathways in mammalian cells to illustrate mechanisms and regulation of deadenylation-dependent mRNA decay, including decay directed by adenine/uridine-rich elements (AREs) in the 3(') -untranslated region (UTR), the rapid decay mediated by destabilizing elements in protein-coding regions, the surveillance mechanism that detects and degrades nonsense-containing mRNA [i.e., nonsense-mediated decay (NMD)], the decay directed by miRNAs, and the default decay pathway for stable messages. Mammalian mRNA deadenylation involves two consecutive phases mediated by the PAN2-PAN3 and the CCR4-CAF1 complexes, respectively. Decapping takes place after deadenylation and may serve as a backup mechanism to trigger mRNA decay if initial deadenylation is compromised. In addition, we discuss how deadenylation impacts the dynamics of RNA processing bodies (P-bodies), where nontranslatable mRNAs can be degraded or stored. Possible models for mechanisms of various deadenylation-dependent mRNA decay pathways are also discussed.
Copyright © 2010 John Wiley & Sons, Ltd.
Figures



References
-
- Wickens M, et al. Translational control in developmental decisions. In: Mathews M, editor. Translational Control. 2. Cold Spring Harbor Press; 2000.
-
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. - PubMed
-
- Garneau NL, et al. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. - PubMed
-
- Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

