Recognition of S-adenosylmethionine by riboswitches
- PMID: 21957011
- PMCID: PMC3618691
- DOI: 10.1002/wrna.63
Recognition of S-adenosylmethionine by riboswitches
Abstract
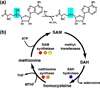
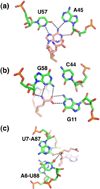
Riboswitches are regulatory elements commonly found in the 5' leader sequences of bacterial mRNAs that bind cellular metabolites to direct expression at either the transcriptional or translational level. The effectors of these RNAs are chemically diverse, including nucleobases and nucleosides, amino acids, cofactors, and second messenger molecules. Over the last few years, a number of structures have revealed the architectural means by which RNA creates binding pockets of high affinity and specificity for these compounds. For most effectors, there is a single class of associated riboswitches. However, eight individual classes of S-adenosylmethionine (SAM) and/or S-adenosylhomocysteine (SAH) responsive riboswitches that control various aspects of sulfur metabolism have been validated, revealing a diverse set of solutions to the recognition of these ubiquitous metabolites. This review focuses upon the structures of RNAs that bind SAM and SAH and how they discriminate between these compounds.
Copyright © 2011 John Wiley & Sons, Ltd.
Figures
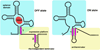
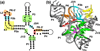
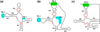
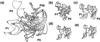
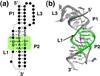
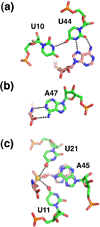
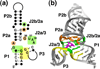
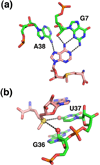
References
-
- Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. - PubMed
-
- Fontecave M, Atta M, Mulliez E. S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci. 2004;29:243–249. - PubMed
-
- Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. - PubMed
-
- Sekowska A, Kung HF, Danchin A. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol. 2000;2:145–177. - PubMed
-
- Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli. Mol Microbiol. 1991;5:1593–1597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

