Mapping QTLs for oil traits and eQTLs for oleosin genes in jatropha
- PMID: 21958236
- PMCID: PMC3195091
- DOI: 10.1186/1471-2229-11-132
Mapping QTLs for oil traits and eQTLs for oleosin genes in jatropha
Abstract
Background: The major fatty acids in seed oil of jatropha, a biofuel crop, are palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1) and linoleic acid (C18:2). High oleic acid and total oil content are desirable for jatropha breeding. Until now, little was known about the genetic bases of these oil traits in jatropha. In this study, quantitative trait locus (QTL) and expression QTL analyses were applied to identify genetic factors that are relevant to seed oil traits in jatropha.
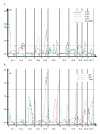
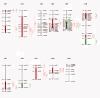
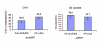
Results: Composite interval mapping identified 18 QTL underlying the oil traits. A highly significant QTL qC18:1-1 was detected at one end of linkage group (LG) 1 with logarithm of the odd (LOD) 18.4 and percentage of variance explained (PVE) 36.0%. Interestingly, the QTL qC18:1-1 overlapped with qC18:2-1, controlling oleic acid and linoleic acid compositions. Among the significant QTL controlling total oil content, qOilC-4 was mapped on LG4 a relatively high significant level with LOD 5.0 and PVE 11.1%. Meanwhile, oleosins are the major composition in oil body affecting oil traits; we therefore developed SNP markers in three oleosin genes OleI, OleII and OleIII, which were mapped onto the linkage map. OleI and OleIII were mapped on LG5, closing to QTLs controlling oleic acid and stearic acid. We further determined the expressions of OleI, OleII and OleIII in mature seeds from the QTL mapping population, and detected expression QTLs (eQTLs) of the three genes on LGs 5, 6 and 8 respectively. The eQTL of OleIII, qOleIII-5, was detected on LG5 with PVE 11.7% and overlapped with QTLs controlling stearic acid and oleic acid, implying a cis- or trans-element for the OleIII affecting fatty acid compositions.
Conclusion: We identified 18 QTLs underlying the oil traits and 3 eQTLs of the oleosin acid genes. The QTLs and eQTLs, especially qC18:1-1, qOilC-4 and qOleIII-5 with contribution rates (R2) higher than 10%, controlling oleic acid, total oil content and oleosin gene expression respectively, will provide indispensable data for initiating molecular breeding to improve seed oil traits in jatropha, the key crop for biodiesel production.
Figures
References
-
- Falconer DS, Mackay TFC, Frankham R. Trends in Genetics. 4. 7. Vol. 12. 1996. Introduction to Quantitative Genetics; p. 280. - DOI
-
- Varshney RK, Tuberosa R. Genomics-assisted Crop Improvement: Genomics applications in crops. Vol. 2. Springer; 2007.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

