NetSlim: high-confidence curated signaling maps
- PMID: 21959865
- PMCID: PMC3263596
- DOI: 10.1093/database/bar032
NetSlim: high-confidence curated signaling maps
Abstract
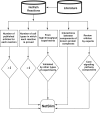
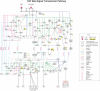
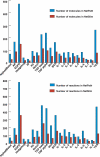
We previously developed NetPath as a resource for comprehensive manually curated signal transduction pathways. The pathways in NetPath contain a large number of molecules and reactions which can sometimes be difficult to visualize or interpret given their complexity. To overcome this potential limitation, we have developed a set of more stringent curation and inclusion criteria for pathway reactions to generate high-confidence signaling maps. NetSlim is a new resource that contains this 'core' subset of reactions for each pathway for easy visualization and manipulation. The pathways in NetSlim are freely available at http://www.netpath.org/netslim.
Figures

References
-
- Saraiya P, North C, Duca K. Visualizing biological pathways: requirements analysis, systems evaluation and research agenda. Inform. Visual. 2005b;4:1–15.
-
- Albert R, Wang RS. Discrete dynamic modeling of cellular signaling networks. Methods Enzymol. 2009;467:281–306. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

