Tripal: a construction toolkit for online genome databases
- PMID: 21959868
- PMCID: PMC3263599
- DOI: 10.1093/database/bar044
Tripal: a construction toolkit for online genome databases
Abstract
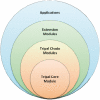
As the availability, affordability and magnitude of genomics and genetics research increases so does the need to provide online access to resulting data and analyses. Availability of a tailored online database is the desire for many investigators or research communities; however, managing the Information Technology infrastructure needed to create such a database can be an undesired distraction from primary research or potentially cost prohibitive. Tripal provides simplified site development by merging the power of Drupal, a popular web Content Management System with that of Chado, a community-derived database schema for storage of genomic, genetic and other related biological data. Tripal provides an interface that extends the content management features of Drupal to the data housed in Chado. Furthermore, Tripal provides a web-based Chado installer, genomic data loaders, web-based editing of data for organisms, genomic features, biological libraries, controlled vocabularies and stock collections. Also available are Tripal extensions that support loading and visualizations of NCBI BLAST, InterPro, Kyoto Encyclopedia of Genes and Genomes and Gene Ontology analyses, as well as an extension that provides integration of Tripal with GBrowse, a popular GMOD tool. An Application Programming Interface is available to allow creation of custom extensions by site developers, and the look-and-feel of the site is completely customizable through Drupal-based PHP template files. Addition of non-biological content and user-management is afforded through Drupal. Tripal is an open source and freely available software package found at http://tripal.sourceforge.net.
Figures
References
-
- Drysdale R. FlyBase : a database for the Drosophila research community. Methods Mol. Biol. 2008;420:45–59. - PubMed
-
- Hirschman JE, Engel S, Hong E, et al. The Saccharomyces Genome Database provides comprehensive information about the biology of S-cerevisiae and tools for studies in comparative genomics. FASEB J. 2007;21:A264–A264.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

