Proteomic analysis of sera from common variable immunodeficiency patients undergoing replacement intravenous immunoglobulin therapy
- PMID: 21960740
- PMCID: PMC3180879
- DOI: 10.1155/2011/706746
Proteomic analysis of sera from common variable immunodeficiency patients undergoing replacement intravenous immunoglobulin therapy
Abstract
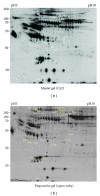
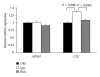
Common variable immunodeficiency is the most common form of symptomatic primary antibody failure in adults and children. Replacement immunoglobulin is the standard treatment of these patients. By using a differential proteomic approach based on 2D-DIGE, we examined serum samples from normal donors and from matched, naive, and immunoglobulin-treated patients. The results highlighted regulated expression of serum proteins in naive patients. Among the identified proteins, clusterin/ApoJ serum levels were lower in naive patients, compared to normal subjects. This finding was validated in a wider collection of samples from newly enrolled patients. The establishment of a cellular system, based on a human hepatocyte cell line HuH7, allowed to ascertain a potential role in the regulation of CLU gene expression by immunoglobulins.
Figures

References
-
- Rosen FS, Eibl M, Roifman C, et al. Primary immunodeficiency diseases. Clinical and Experimental Immunology, Supplement. 1999;118(1):1–28.
-
- Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clinical Immunology. 1999;92(1):34–48. - PubMed
-
- Park MA, Li JT, Hagan JB, Maddox DE, Abraham RS. Common variable immunodeficiency: a new look at an old disease. The Lancet. 2008;372(9637):489–502. - PubMed
-
- Ardeniz O, Başoğlu OK, Günşar F, et al. Clinical and immunological analysis of 23 adult patients with common variable immunodeficiency. Journal of Investigational Allergology and Clinical Immunology. 2010;20(3):222–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

