Bound water at protein-protein interfaces: partners, roles and hydrophobic bubbles as a conserved motif
- PMID: 21961043
- PMCID: PMC3178540
- DOI: 10.1371/journal.pone.0024712
Bound water at protein-protein interfaces: partners, roles and hydrophobic bubbles as a conserved motif
Abstract
Background: There is a great interest in understanding and exploiting protein-protein associations as new routes for treating human disease. However, these associations are difficult to structurally characterize or model although the number of X-ray structures for protein-protein complexes is expanding. One feature of these complexes that has received little attention is the role of water molecules in the interfacial region.
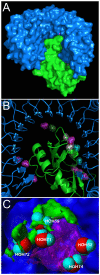
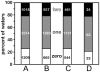
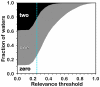
Methodology: A data set of 4741 water molecules abstracted from 179 high-resolution (≤ 2.30 Å) X-ray crystal structures of protein-protein complexes was analyzed with a suite of modeling tools based on the HINT forcefield and hydrogen-bonding geometry. A metric termed Relevance was used to classify the general roles of the water molecules.
Results: The water molecules were found to be involved in: a) (bridging) interactions with both proteins (21%), b) favorable interactions with only one protein (53%), and c) no interactions with either protein (26%). This trend is shown to be independent of the crystallographic resolution. Interactions with residue backbones are consistent for all classes and account for 21.5% of all interactions. Interactions with polar residues are significantly more common for the first group and interactions with non-polar residues dominate the last group. Waters interacting with both proteins stabilize on average the proteins' interaction (-0.46 kcal mol(-1)), but the overall average contribution of a single water to the protein-protein interaction energy is unfavorable (+0.03 kcal mol(-1)). Analysis of the waters without favorable interactions with either protein suggests that this is a conserved phenomenon: 42% of these waters have SASA ≤ 10 Å(2) and are thus largely buried, and 69% of these are within predominantly hydrophobic environments or "hydrophobic bubbles". Such water molecules may have an important biological purpose in mediating protein-protein interactions.
Conflict of interest statement
Figures





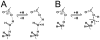


References
-
- Zacharias M. 400 p; 2010. Protein-protein complexes: Analysis, modeling and drug design: Imperial College Press.
-
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. - PubMed
-
- Wilson AJ. Inhibition of protein-protein interactions using designed molecules. Chem Soc Rev. 2009;38:3289–3300. - PubMed
-
- Betzi S, Guerlesquin F, Morelli X. Protein-protein interaction inhibition (2P2I): fewer and fewer undruggable targets. Comb Chem High Throughput Screen. 2009;12:968–983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

