Site-selective artificial ribonucleases: oligonucleotide conjugates containing multiple imidazole residues in the catalytic domain
- PMID: 21961054
- PMCID: PMC3180074
- DOI: 10.4061/2011/748632
Site-selective artificial ribonucleases: oligonucleotide conjugates containing multiple imidazole residues in the catalytic domain
Abstract
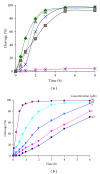
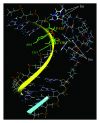
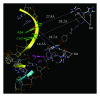
Design of site-selective artificial ribonucleases (aRNases) is one of the most challenging tasks in RNA targeting. Here, we designed and studied oligonucleotide-based aRNases containing multiple imidazole residues in the catalytic part and systematically varied structure of cleaving constructs. We demonstrated that the ribonuclease activity of the conjugates is strongly affected by the number of imidazole residues in the catalytic part, the length of a linker between the catalytic imidazole groups of the construct and the oligonucleotide, and the type of anchor group, connecting linker structure and the oligonucleotide. Molecular modeling of the most active aRNases showed that preferable orientation(s) of cleaving constructs strongly depend on the structure of the anchor group and length of the linker. The inclusion of deoxyribothymidine anchor group significantly reduced the probability of cleaving groups to locate near the cleavage site, presumably due to a stacking interaction with the neighbouring nucleotide residue. Altogether the obtained results show that dynamics factors play an important role in site-specific RNA cleavage. Remarkably high cleavage activity was displayed by the conjugates with the most flexible and extended cleaving construct, which presumably provides a better opportunity for imidazole residues to be correctly positioned in the vicinity of scissile phosphodiester bond.
Figures






References
-
- Zenkova MA, Beloglazova NG. Site-specific artificial ribonucleases: conjugates of oligonucleotides with catalytic groups. In: Zenkova MA, editor. Nucleic Acids and Molecular Biology: Artificial Nucleases. Vol. 13. Berlin, Germany: Springer; 2004. pp. 189–222.
-
- Sil'nikov VN, Vlassov VV. Design of site-specific RNA-cleaving reagents. Russian Chemical Reviews. 2001;70(6):491–508.
-
- Lönnberg H. Cleavage of RNA phosphodiester bonds by small molecular entities: a mechanistic insight. Organic and Biomolecular Chemistry. 2011;21:1687–1703. - PubMed
-
- Grimm GN, Boutorine AS, Hélène C. Rapid routes of synthesis of oligonucleotide conjugates from non-protected oligonucleotides and ligands possessing different nucleophilic or electrophilic functional groups. Nucleosides, Nucleotides and Nucleic Acids. 2000;19(10–12):1943–1965. - PubMed
-
- Modak AS, Gard JK, Merriman MC, Winkeler KA, Bashkin JK, Stern MK. Toward chemical ribonucleases 2. Synthesis and characterization of nucleoside-bipyridine conjugates. Hydrolytic cleavage of RNA by their copper(II) complexes. Journal of the American Chemical Society. 1991;113(1):283–291.
LinkOut - more resources
Full Text Sources

