Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153
- PMID: 21961597
- PMCID: PMC3183753
- DOI: 10.1016/j.bpj.2011.08.025
Single molecule study of the intrinsically disordered FG-repeat nucleoporin 153
Abstract
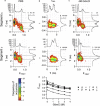
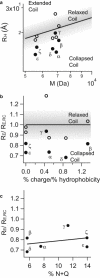
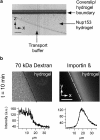
Nucleoporins (Nups), which are intrinsically disordered, form a selectivity filter inside the nuclear pore complex, taking a central role in the vital nucleocytoplasmic transport mechanism. These Nups display a complex and nonrandom amino-acid architecture of phenylalanine glycine (FG)-repeat clusters and intra-FG linkers. How such heterogeneous sequence composition relates to function and could give rise to a transport mechanism is still unclear. Here we describe a combined chemical biology and single-molecule fluorescence approach to study the large human Nup153 FG-domain. In order to obtain insights into the properties of this domain beyond the average behavior, we probed the end-to-end distance (R(E)) of several ∼50-residues long FG-repeat clusters in the context of the whole protein domain. Despite the sequence heterogeneity of these FG-clusters, we detected a reoccurring and consistent compaction from a relaxed coil behavior under denaturing conditions (R(E)/R(E,RC) = 0.99 ± 0.15 with R(E,RC) corresponding to ideal relaxed coil behavior) to a collapsed state under native conditions (R(E)/R(E,RC) = 0.79 ± 0.09). We then analyzed the properties of this protein on the supramolecular level, and determined that this human FG-domain was in fact able to form a hydrogel with physiological permeability barrier properties.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

