Clan genomics and the complex architecture of human disease
- PMID: 21962505
- PMCID: PMC3656718
- DOI: 10.1016/j.cell.2011.09.008
Clan genomics and the complex architecture of human disease
Abstract
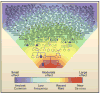
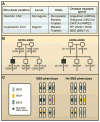
Human diseases are caused by alleles that encompass the full range of variant types, from single-nucleotide changes to copy-number variants, and these variations span a broad frequency spectrum, from the very rare to the common. The picture emerging from analysis of whole-genome sequences, the 1000 Genomes Project pilot studies, and targeted genomic sequencing derived from very large sample sizes reveals an abundance of rare and private variants. One implication of this realization is that recent mutation may have a greater influence on disease susceptibility or protection than is conferred by variations that arose in distant ancestors.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. - PubMed
-
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997a;277:1805–1807. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

