Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes
- PMID: 21962510
- PMCID: PMC3190307
- DOI: 10.1016/j.cell.2011.07.048
Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes
Abstract
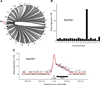
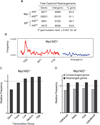
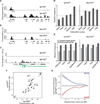
Chromosomal rearrangements, including translocations, require formation and joining of DNA double strand breaks (DSBs). These events disrupt the integrity of the genome and are frequently involved in producing leukemias, lymphomas and sarcomas. Despite the importance of these events, current understanding of their genesis is limited. To examine the origins of chromosomal rearrangements we developed Translocation Capture Sequencing (TC-Seq), a method to document chromosomal rearrangements genome-wide, in primary cells. We examined over 180,000 rearrangements obtained from 400 million B lymphocytes, revealing that proximity between DSBs, transcriptional activity and chromosome territories are key determinants of genome rearrangement. Specifically, rearrangements tend to occur in cis and to transcribed genes. Finally, we find that activation-induced cytidine deaminase (AID) induces the rearrangement of many genes found as translocation partners in mature B cell lymphoma.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Translocation mapping exposes the risky lifestyle of B cells.Cell. 2011 Sep 30;147(1):20-2. doi: 10.1016/j.cell.2011.09.005. Cell. 2011. PMID: 21962501
-
Cancer genomics: Translocation patterns revealed.Nat Rev Genet. 2011 Oct 18;12(11):741. doi: 10.1038/nrg3090. Nat Rev Genet. 2011. PMID: 22005978 No abstract available.
References
-
- Argast GM, Stephens KM, Emond MJ, Monnat RJ., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J Mol Biol. 1998;280:345–353. - PubMed
-
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

