A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells
- PMID: 21962512
- PMCID: PMC3216680
- DOI: 10.1016/j.cell.2011.08.038
A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells
Abstract
The transcriptional activators Oct4, Sox2, and Nanog cooperate with a wide array of cofactors to orchestrate an embryonic stem (ES) cell-specific gene expression program that forms the molecular basis of pluripotency. Here, we report using an unbiased in vitro transcription-biochemical complementation assay to discover a multisubunit stem cell coactivator complex (SCC) that is selectively required for the synergistic activation of the Nanog gene by Oct4 and Sox2. Purification, identification, and reconstitution of SCC revealed this coactivator to be the trimeric XPC-nucleotide excision repair complex. SCC interacts directly with Oct4 and Sox2 and is recruited to the Nanog and Oct4 promoters as well as a majority of genomic regions that are occupied by Oct4 and Sox2. Depletion of SCC/XPC compromised both pluripotency in ES cells and somatic cell reprogramming of fibroblasts to induced pluripotent stem (iPS) cells. This study identifies a transcriptional coactivator with diversified functions in maintaining ES cell pluripotency and safeguarding genome integrity.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


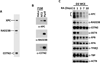

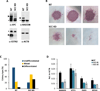
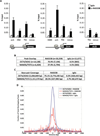

Comment in
-
eNERgizing pluripotent gene transcription.Cell Stem Cell. 2011 Oct 4;9(4):285-6. doi: 10.1016/j.stem.2011.09.004. Cell Stem Cell. 2011. PMID: 21982224
References
-
- Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001;276:18665–18672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

