Long-range massively parallel mate pair sequencing detects distinct mutations and similar patterns of structural mutability in two breast cancer cell lines
- PMID: 21962895
- PMCID: PMC3185296
- DOI: 10.1016/j.cancergen.2011.07.009
Long-range massively parallel mate pair sequencing detects distinct mutations and similar patterns of structural mutability in two breast cancer cell lines
Erratum in
- Cancer Genet. 2011 Dec;204(12):694. Coarfa, Cristian [added];Schoenherr, Caroline [added]
Abstract
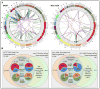
Cancer genomes frequently undergo genomic instability resulting in accumulation of chromosomal rearrangement. To date, one of the main challenges has been to confidently and accurately identify these rearrangements by using short-read massively parallel sequencing. We were able to improve cancer rearrangement detection by combining two distinct massively parallel sequencing strategies: fosmid-sized (36 kb on average) and standard 5 kb mate pair libraries. We applied this combined strategy to map rearrangements in two breast cancer cell lines, MCF7 and HCC1954. We detected and validated a total of 91 somatic rearrangements in MCF7 and 25 in HCC1954, including genomic alterations corresponding to previously reported transcript aberrations in these two cell lines. Each of the genomes contains two types of breakpoints: clustered and dispersed. In both cell lines, the dispersed breakpoints show enrichment for low copy repeats, while the clustered breakpoints associate with high copy number amplifications. Comparing the two genomes, we observed highly similar structural mutational spectra affecting different sets of genes, pointing to similar histories of genomic instability against the background of very different gene network perturbations.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

References
-
- Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, Campbell P, Quail M, Plumb B, Matthews L, McLay K, Edwards PA, Rogers J, Wooster R, Futreal PA, Stratton MR. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–303. - PMC - PubMed
-
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. - PMC - PubMed
-
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, Olson MV, Eichler EE. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. - PubMed
-
- Volik S, Raphael BJ, Huang G, Stratton MR, Bignel G, Murnane J, Brebner JH, Bajsarowicz K, Paris PL, Tao Q, Kowbel D, Lapuk A, Shagin DA, Shagina IA, Gray JW, Cheng JF, de Jong PJ, Pevzner P, Collins C. Decoding the fine-scale structure of a breast cancer genome and transcriptome. Genome Res. 2006 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

