A conserved regulatory role for antisense RNA in meiotic gene expression in yeast
- PMID: 21963111
- PMCID: PMC3230709
- DOI: 10.1016/j.mib.2011.09.010
A conserved regulatory role for antisense RNA in meiotic gene expression in yeast
Abstract
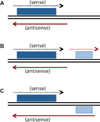
A significant fraction of the eukaryotic genome is transcribed into RNAs that do not encode proteins, termed non-coding RNA (ncRNA). One class of ncRNA that is of particular interest is antisense RNAs, which are complementary to protein coding transcripts (mRNAs). In this article, we summarize recent studies using different yeasts that reveal a conserved pattern in which meiotically expressed genes have antisense transcripts in vegetative cells. These antisense transcripts repress the basal transcription of the mRNA during vegetative growth and are diminished as cells enter meiosis. While the mechanism(s) by which these antisense RNAs interfere with production of sense transcripts is not yet understood, the effects appear to be independent of the canonical RNAi machinery.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
-
- Nakamura T, Kishida M, Shimoda C. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells. 2000;5:463–479. - PubMed
-
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. - PubMed
-
-
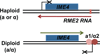
Gelfand B, Mead J, Bruning A, Apostolopoulos N, Tadigotla V, Nagaraj V, Sengupta AM, Vershon AK. Regulated antisense transcription controls expression of cell-type specific genes in yeast. Mol Cell Biol. 2011;31:1701–1709. Analysis of the sense/antisense pair IME4/RME2 indicates that RME2 interferes with IME4 expression by inhibiting elongation rather than inititation of the sense transcript.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

