Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis
- PMID: 21963668
- PMCID: PMC3203429
- DOI: 10.1105/tpc.111.088583
Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis
Abstract
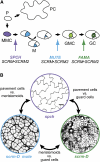
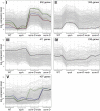
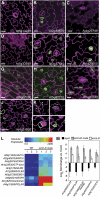
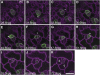
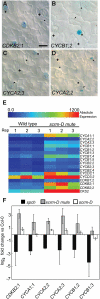
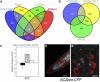
The balance between maintenance and differentiation of stem cells is a central question in developmental biology. Development of stomata in Arabidopsis thaliana begins with de novo asymmetric divisions producing meristemoids, proliferating precursor cells with stem cell-like properties. The transient and asynchronous nature of the meristemoid has made it difficult to study its molecular characteristics. Synthetic combination of stomatal differentiation mutants due to loss- or gain-of-function mutations in SPEECHLESS, MUTE, and SCREAM create seedlings with an epidermis overwhelmingly composed of pavement cells, meristemoids, or stomata, respectively. Through transcriptome analysis, we define and characterize the molecular signatures of meristemoids. The reporter localization studies of meristemoid-enriched proteins reveals pathways not previously associated with stomatal development. We identified a novel protein, POLAR, and demonstrate through time-lapse live imaging that it exhibits transient polar localization and segregates unevenly during meristemoid asymmetric divisions. The polar localization of POLAR requires BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE. Comparative bioinformatic analysis of the transcriptional profiles of a meristemoid with shoot and root apical meristems highlighted cytokinin signaling and the ERECTA family receptor-like kinases in the broad regulation of stem cell populations. Our work reveals molecular constituents of stomatal stem cells and illuminates a common theme among stem cell populations in plants.
Figures


References
-
- Abrash E.B., Bergmann D.C. (2009). Asymmetric cell divisions: A view from plant development. Dev. Cell 16: 783–796 - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300
-
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 - PubMed
-
- Bergmann D.C., Sack F.D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58: 163–181 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

