Molecular mechanism of transcription inhibition by phage T7 gp2 protein
- PMID: 21963987
- PMCID: PMC3217721
- DOI: 10.1016/j.jmb.2011.09.029
Molecular mechanism of transcription inhibition by phage T7 gp2 protein
Abstract
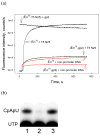
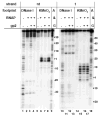
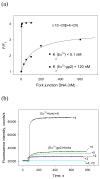
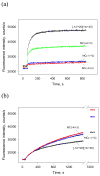
Escherichia coli T7 bacteriophage gp2 protein is a potent inhibitor of host RNA polymerase (RNAP). gp2 inhibits formation of open promoter complex by binding to the β' jaw, an RNAP domain that interacts with downstream promoter DNA. Here, we used an engineered promoter with an optimized sequence to obtain and characterize a specific promoter complex containing RNAP and gp2. In this complex, localized melting of promoter DNA is initiated but does not propagate to include the point of the transcription start. As a result, the complex is transcriptionally inactive. Using a highly sensitive RNAP beacon assay, we performed quantitative real-time measurements of specific binding of the RNAP-gp2 complex to promoter DNA and various promoter fragments. In this way, the effect of gp2 on RNAP interaction with promoters was dissected. As expected, gp2 greatly decreased RNAP affinity to downstream promoter duplex. However, gp2 also inhibited RNAP binding to promoter fragments that lacked downstream promoter DNA that interacts with the β' jaw. The inhibition was caused by gp2-mediated decrease of the RNAP binding affinity to template and non-template strand segments of the transcription bubble downstream of the -10 promoter element. The inhibition of RNAP interactions with single-stranded segments of the transcription bubble by gp2 is a novel effect, which may occur via allosteric mechanism that is set in motion by the gp2 binding to the β' jaw.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



References
-
- Nechaev S, Severinov K. Bacteriophage-induced modifications of host RNA polymerase. Annu Rev Microbiol. 2003;57:301–322. - PubMed
-
- Molineux IJ. The T7 group. In: Calendar RL, editor. The Bacteriophages. 2. Oxford University Press; New York: 2005. pp. 277–301.
-
- Hessellbach B, Nakada D. Inactive complex formation between E. coli RNA polymerase and inhibitor protein purified from T7 phage infected cells. Nature. 1975;258:354–357. - PubMed
-
- Nechaev S, Severinov K. Inhibition of Escherichia coli RNA polymerase by bacteriophage T7 gene 2 protein. J Mol Biol. 1999;289:815–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

