miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA
- PMID: 21964070
- PMCID: PMC3230379
- DOI: 10.1038/emboj.2011.359
miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA
Abstract
MicroRNAs (miRNAs) are ∼22 nt non-coding RNAs that typically bind to the 3' UTR of target mRNAs in the cytoplasm, resulting in mRNA destabilization and translational repression. Here, we report that miRNAs can also regulate gene expression by targeting non-coding antisense transcripts in human cells. Specifically, we show that miR-671 directs cleavage of a circular antisense transcript of the Cerebellar Degeneration-Related protein 1 (CDR1) locus in an Ago2-slicer-dependent manner. The resulting downregulation of circular antisense has a concomitant decrease in CDR1 mRNA levels, independently of heterochromatin formation. This study provides the first evidence for non-coding antisense transcripts as functional miRNA targets, and a novel regulatory mechanism involving a positive correlation between mRNA and antisense circular RNA levels.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


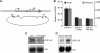

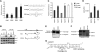
Comment in
-
A novel nuclear miRNA mediated modulation of a non-coding antisense RNA and its cognate sense coding mRNA.EMBO J. 2011 Nov 2;30(21):4340-1. doi: 10.1038/emboj.2011.373. EMBO J. 2011. PMID: 22048334 Free PMC article.
References
-
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73: 1019–1030 - PubMed
-
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447: 823–828 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

