Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding
- PMID: 21964413
- PMCID: PMC3196379
- DOI: 10.1038/nbt.1977
Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding
Abstract
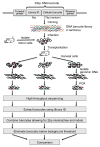
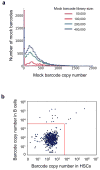
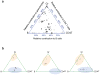
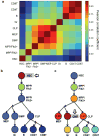
Disentangling cellular heterogeneity is a challenge in many fields, particularly in the stem cell and cancer biology fields. Here we demonstrate how to combine viral genetic barcoding with high-throughput sequencing to track single cells in a heterogeneous population. We use this technique to track the in vivo differentiation of unitary hematopoietic stem cells (HSCs). The results are consistent with single-cell transplantation studies but require two orders of magnitude fewer mice. In addition to its high throughput, the high sensitivity of the technique allows for a direct examination of the clonality of sparse cell populations such as HSCs. We show how these capabilities offer a clonal perspective of the HSC differentiation process. In particular, our data suggest that HSCs do not equally contribute to blood cells after irradiation-mediated transplantation, and that two distinct HSC differentiation patterns co-exist in the same recipient mouse after irradiation. This technique can be applied to any virus-accessible cell type for both in vitro and in vivo processes.
Figures

Comment in
-
You can count on this: barcoded hematopoietic stem cells.Cell Stem Cell. 2011 Nov 4;9(5):390-2. doi: 10.1016/j.stem.2011.10.013. Cell Stem Cell. 2011. PMID: 22056135
-
One stem cell at a time.Nat Methods. 2011 Nov;8(11):896-7. doi: 10.1038/nmeth.1752. Nat Methods. 2011. PMID: 22148155 No abstract available.
References
-
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. S0092-8674(00)81692-X [pii] - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

