Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome
- PMID: 21964572
- PMCID: PMC3235474
- DOI: 10.1038/ng.944
Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome
Abstract
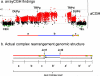
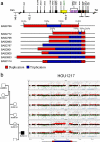
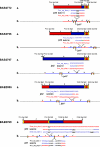
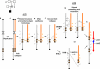
We identified complex genomic rearrangements consisting of intermixed duplications and triplications of genomic segments at the MECP2 and PLP1 loci. These complex rearrangements were characterized by a triplicated segment embedded within a duplication in 11 unrelated subjects. Notably, only two breakpoint junctions were generated during each rearrangement formation. All the complex rearrangement products share a common genomic organization, duplication-inverted triplication-duplication (DUP-TRP/INV-DUP), in which the triplicated segment is inverted and located between directly oriented duplicated genomic segments. We provide evidence that the DUP-TRP/INV-DUP structures are mediated by inverted repeats that can be separated by >300 kb, a genomic architecture that apparently leads to susceptibility to such complex rearrangements. A similar inverted repeat-mediated mechanism may underlie structural variation in many other regions of the human genome. We propose a mechanism that involves both homology-driven events, via inverted repeats, and microhomologous or nonhomologous events.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

