Gadd45β is an inducible coactivator of transcription that facilitates rapid liver growth in mice
- PMID: 21965327
- PMCID: PMC3204825
- DOI: 10.1172/JCI38760
Gadd45β is an inducible coactivator of transcription that facilitates rapid liver growth in mice
Abstract
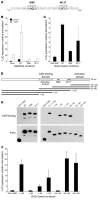
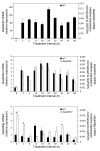
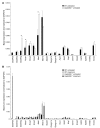
The growth arrest and DNA damage-inducible 45 (Gadd45) proteins act in many cellular processes. In the liver, Gadd45b (encoding Gadd45β) is the gene most strongly induced early during both compensatory regeneration and drug-induced hyperplasia. The latter response is associated with the dramatic and rapid hepatocyte growth that follows administration of the xenobiotic TCPOBOP (1,4-bis[2-(3,5)-dichoropyridyloxy] benzene), a ligand of the nuclear receptor constitutive androstane receptor (CAR). Here, we have shown that Gadd45b-/- mice have intact proliferative responses following administration of a single dose of TCPOBOP, but marked growth delays. Moreover, early transcriptional stimulation of CAR target genes was weaker in Gadd45b-/- mice than in wild-type animals, and more genes were downregulated. Gadd45β was then found to have a direct role in transcription by physically binding to CAR, and TCPOBOP treatment caused both proteins to localize to a regulatory element for the CAR target gene cytochrome P450 2b10 (Cyp2b10). Further analysis defined separate Gadd45β domains that mediated binding to CAR and transcriptional activation. Although baseline hepatic expression of Gadd45b was broadly comparable to that of other coactivators, its 140-fold stimulation by TCPOBOP was striking and unique. The induction of Gadd45β is therefore a response that facilitates increased transcription, allowing rapid expansion of liver mass for protection against xenobiotic insults.
Figures

References
-
- Ledda-Columbano GM, Pibiri M, Loi R, Perra A, Shinozuka H, Columbano A. Early increase in cyclin-D1 expression and accelerated entry of mouse hepatocytes into S phase after administration of the mitogen 1, 4-Bis[2- (3,5-Dichloropyridyloxy)] benzene. Am J Pathol. 2000;156(1):91–97. doi: 10.1016/S0002-9440(10)64709-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

