High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes
- PMID: 21965408
- PMCID: PMC3233045
- DOI: 10.1128/AEM.00646-11
High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes
Abstract
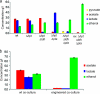
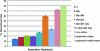
This work describes novel genetic tools for use in Clostridium thermocellum that allow creation of unmarked mutations while using a replicating plasmid. The strategy employed counter-selections developed from the native C. thermocellum hpt gene and the Thermoanaerobacterium saccharolyticum tdk gene and was used to delete the genes for both lactate dehydrogenase (Ldh) and phosphotransacetylase (Pta). The Δldh Δpta mutant was evolved for 2,000 h, resulting in a stable strain with 40:1 ethanol selectivity and a 4.2-fold increase in ethanol yield over the wild-type strain. Ethanol production from cellulose was investigated with an engineered coculture of organic acid-deficient engineered strains of both C. thermocellum and T. saccharolyticum. Fermentation of 92 g/liter Avicel by this coculture resulted in 38 g/liter ethanol, with acetic and lactic acids below detection limits, in 146 h. These results demonstrate that ethanol production by thermophilic, cellulolytic microbes is amenable to substantial improvement by metabolic engineering.
Figures
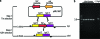
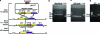

References
-
- Alper H., Stephanopoulos G. 2009. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat. Rev. Microbiol. 7: 715–723 - PubMed
-
- Bayer E. A., Lamed R., White B. A., Flint H. J. 2008. From cellulosomes to cellulosomics. Chem. Rec. 8: 364–377 - PubMed
-
- Benoit L., Cailliez C., Petitdemange E., Gitton J. 1992. Isolation of cellulolytic mesophilic clostridia from a municipal solid waste digestor. Microb. Ecol. 23: 117–125 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

