Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism
- PMID: 21965561
- PMCID: PMC3232889
- DOI: 10.1128/JB.05987-11
Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism
Abstract
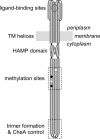
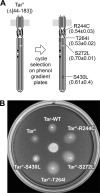
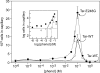
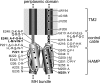
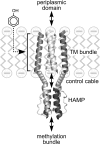
The four transmembrane chemoreceptors of Escherichia coli sense phenol as either an attractant (Tar) or a repellent (Tap, Trg, and Tsr). In this study, we investigated the Tar determinants that mediate its attractant response to phenol and the Tsr determinants that mediate its repellent response to phenol. Tar molecules with lesions in the aspartate-binding pocket of the periplasmic domain, with a foreign periplasmic domain (from Tsr or from several Pseudomonas chemoreceptors), or lacking nearly the entire periplasmic domain still mediated attractant responses to phenol. Similarly, Tar molecules with the cytoplasmic methylation and kinase control domains of Tsr still sensed phenol as an attractant. Additional hybrid receptors with signaling elements from both Tar and Tsr indicated that the transmembrane (TM) helices and HAMP domain determined the sign of the phenol-sensing response. Several amino acid replacements in the HAMP domain of Tsr, particularly attractant-mimic signaling lesions at residue E248, converted Tsr to an attractant sensor of phenol. These findings suggest that phenol may elicit chemotactic responses by diffusing into the cytoplasmic membrane and perturbing the structural stability or position of the TM bundle helices, in conjunction with structural input from the HAMP domain. We conclude that behavioral responses to phenol, and perhaps to temperature, cytoplasmic pH, and glycerol, as well, occur through a general sensing mechanism in chemoreceptors that detects changes in the structural stability or dynamic behavior of a receptor signaling element. The structurally sensitive target for phenol is probably the TM bundle, but other behaviors could target other receptor elements.
Figures



References
-
- Adler J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77–91 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

